Abstract
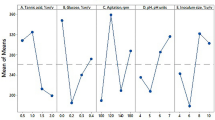
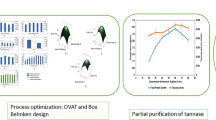
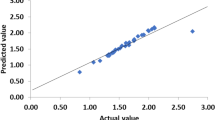
The optimization of tannase production by Lactobacillus plantarum CIR1 was carried out following the Taguchi methodology. The orthogonal array employed was L18 (21 × 35) considering six important factors (pH and temperature, also phosphate, nitrogen, magnesium, and carbon sources) for tannase biosynthesis. The experimental results obtained from 18 trials were processed using the software Statistical version 7.1 using the character higher the better. Optimal culture conditions were pH, 6; temperature, 40 °C; tannic acid, 15.0 g/L; KH2PO4, 1.5 g/L; NH4Cl, 7.0 g/L; and MgSO4, 1.5 g/L which were obtained and further validated resulting in an enhance tannase yield of 2.52-fold compared with unoptimized conditions. Tannase production was further carried out in a 1-L gas-lift bioreactor where two nitrogen flows (0.5 and 1.0 vvm) were used to provide anaerobic conditions. Taguchi methodology allowed obtaining the optimal culture conditions for the production of tannase by L. plantarum CIR1. At the gas-lift bioreactor the tannase productivity yields increase 5.17 and 8.08-fold for the flow rates of 0.5 and 1.0 vvm, respectively. Lactobacillus plantarum CIR1 has the capability to produce tannase at laboratory-scale. This is the first report for bacterial tannase production using a gas-lift bioreactor.






Similar content being viewed by others
References
Lekha PK, Lonsane BK (1997) Production and application of tannin acyl hydrolase: state of the art. Adv Appl Microbiol 44:215–260
Aguilar CN, Gutiérrez-Sánchez G (2001) Review sources, properties, applications and potential uses of tannin acyl hydrolase. Food Sci Technol Int 7:373–382
Belmares R, Contreras-Esquivel JC, Rodriguez-Herrera R, Ramirez-Coronel A, Aguilar CN (2004) Microbial production of tannase: an enzyme with potential use in food industry. Lebensm Wiss Technol 37:857–864
Abdelwahed A, Bouhlel I, Skandrani I, Valenti K, Kadri M, Guiraud P, Steiman R, Mariotte AM, Ghedira K, Laporte F, Dijoux-Franca MG, Chekir-Ghedira L (2007) Study of antimutagenic and antioxidant activities of Gallic acid and 1,2,3,4,6-pentagalloylglucose from Pistacia lentiscus confirmation by microarray expression profiling. Chem Biol Interact 165(1):1–13
Banerjee D, Mahapatra S, Pati BR (2007) Gallic acid production by submerged fermentation of Aspergillus aculeatus DBF9. Res J Microbiol 2(5):462–468
Yu XW, Li YQ (2008) Expression of Aspergillus oryzae tannase in Pichia pastoris and its application in the synthesis of propyl gallate in organic solvent. Food Technol Biotechnol 46(1):80–85
Treviño L, Contreras-Esquivel J, Rodriguez-Herrera R, Aguilar C (2007) Effects of polyurethane matrices on fungal tannase and gallic acid production under solid state culture. J Zhejiang Univ Sci B 8(10):771–776
Mondal KC, Banerjee D, Banerjee R, Pati BR (2001) Production and characterization of tannase from Bacillus cereus KBR9. J Gen Appl Microbiol 47:263–267
Rodríguez H, de las Rivas B, Gómez-Cordovés C, Muñoz R (2008) Characteriztaion of tannase activity in cell-free extracts of Lactobacillus plantarum CECT 748. Int J Food Microbiol 121:92–98
Milva P, Lucia RL, Roberto A, Alessandro E, Guido P, Monia R, Arianna L, Antonio F, Simone G, Silvano EF (2010) Tannic acid degradation by bacterial strains Serratia spp. and Pantoea sp. isolated from olive mill waste mixtures. Int Biodeter Biodegr 64:73–80
Chávez-González M, Rodríguez-Duran LV, Balagurusami N, Prado-Barragán A, Rodríguez R, Contreras JC, Aguilar CN (2011) Biotechnological advances and challenges of tannase: an overview. Food Bioprocess Tech 5(2):445–459
Aguilar CN, Rodríguez R, Gutiérrez-Sánchez G, Augur C, Favela-Torres E, Prado-Barragán LA, Ramírez-Coronel A, Contreras-Esquivel JC (2007) Microbial tannases: advances and perspectives. Appl Microbiol Biotechnol 76:47–59
Rao RS, Kumar GC, Prakasham SR, Hobbs PJ (2008) The Taguchi methodology as a statistical tool for biotechnological applications: a critical appraisal. Biotechnol J 3:510–523
Arvidsson M, Gremyr I (2007) Principles of robust design methodology. Qual Reliab Eng Int 24:23–35
Zhou J, Wu D, Guo D (2010) Optimization of the production of thiocarbohydrazide using the Taguchi method. J Chem Technol Biotechnol 85:1402–1406
Phadke MS, Dehnad K (1988) Optimization of product and process design for quality and cost. Qual Reliab Eng Int 4:159–169
Matin KT, Bastani D, Kazemian H (2009) Applying the Taguchi method to develop an optimized synthesis procedure for nanocrystals of T-type zeolite. Chem Eng Technol 32:1042–1048
Aguilar-Zárate P (2013) Definición de las condiciones de producción de una tanasa bacteriana. M.Sc. thesis. Universidad Autónoma de Coahuila, Saltillo, México
Mondal KC, Pati BR (2000) Studies on the extracellular tannase from newly isolated Bacillus licheniformis KBR6. J Basic Microbiol 40:223–232
Das Mohapatra PK, Maity C, Rao RS, Pati BR, Mondal KC (2009) Tannase production by Bacillus licheniformis KBR6: optimization of submerged culture conditions by Taguchi DOE methodology. Food Res Int 42:430–435
Sharma S, Bhat TK, Dawra TK (2000) A spectrophotometric method for assay of tannase using rhodanine. Anal Biochem 279:85–89
Gatto M, Muratory S, Rinaldi S (1988) A functional interpretation of the logistic equation. Ecol Model 42(2):155–159
Aguilar CN, Augur C, Favela-Torres E, Viniegra-González G (2001) Production of tannase by Aspergillus niger Aa-20 in submerged and solid state fermentation: influence of glucose and tannic acid. J Ind Microbiol Biotechnol 26:296–302
Luedeking R, Piret EL (1959) A kinetic study of the lactic acid fermentation. J Biochem Microbiol Technol Eng 1:393–412
Viniegra-González G, Favela-Torres E, Aguilar CN, Romero-Gómez SJ, Díaz-Godínez G, Augur C (2003) Advantages of fungal enzyme production in solid state over liquid fermentation systems. Biochem Eng J 13:157–167
Kar B, Banerjee R, Bhattacharyya BC (2003) Effect of additives on the behavioral properties of tannin acyl hydrolase. Process Biochem 38:1285–1293
Roy RK (2001) Design of experiments using the Taguchi approach: 16 steps to product and process improvement. John Wiley and Sons, New York
Prasad KK, Mohan SV, Rao RS, Pati BR, Sarma PN (2005) Laccase production by Pleurotus ostreatus 1804: optimization of submerged culture conditions by taguchi DOE methodology. Biochem Eng J 24(1):17–26
Kannan N, Aravindan R (2012) Evaluation and optimization of food-grade tannin acyl hydrolase production by a probiotic Lactobacillus plantarum strain in submerged and solid state fermentation. Food Bioprod Process 90:780–792
Belur PD, Mugeraya G, Nirmala KR, Basavaraj N (2010) Production of novel cell-associated tannase from newly isolated Serratia ficaria DTC. J Microbiol Biotechnol 20(4):732–736
Kannan N, Aravindan R, Viruthagiri T (2011) Effect of culture conditions and kinetic studies on extracellular tannase production by Lactobacillus plantarum MTCC 1407. IJBT 10:321–328
Ruiz HA, Rodríguez-Jasso RM, Rodríguez R, Contreras-Esquivel JC, Aguilar CN (2012) Pectinase production from lemon peel pomace as support and carbon source in solid-state fermentation column-tray bioreactor. Biochem Eng J 65:90–95
Porruat H, Regerat F, Porruat A, Jean D (1982) Production of tannase (tannin acyl hidrolase E.C. 3.1.1.20) by a strain of Aspergillus niger. Biotecnol Lett 44:583–588
Deschamps AM, Otuk G, Lebeault JM (1983) Production of tannase and degradation of chestnut tannin by bacteria. J Ferment Technol 61:55–59
Znad H, Báles V, Markos J, Kawase Y (2004) Modelling and simulation of gas-lift bioreactors. Biochem Eng J 21:73–81
Acknowledgments
Author Pedro Aguilar-Zárate appreciates the scholarship provided by the Mexican Council for Science and Technology (CONACYT).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Aguilar-Zarate, P., Cruz-Hernandez, M.A., Montañez, J.C. et al. Enhancement of tannase production by Lactobacillus plantarum CIR1: validation in gas-lift bioreactor. Bioprocess Biosyst Eng 37, 2305–2316 (2014). https://doi.org/10.1007/s00449-014-1208-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-014-1208-3