Abstract
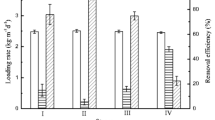
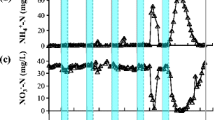
The effect of heavy metals on community structure of a heavy metal tolerant sulfidogenic consortium was evaluated by using a combination of denaturing gradient gel electrophoresis (DGGE) of 16S rRNA gene and dissimilatory sulfite reductase (dsrB) gene fragments, 16S rRNA gene cloning analysis and fluorescence in situ hybridization (FISH). For this purpose, four anaerobic semi-continuous stirred tank reactors (referred as R1–R4) were run in parallel for 12 weeks at heavy metal loading rates of 1.5, 3, 4.5 and 7.5 mg l−1 d−1 each of Cu2+, Ni2+, Zn2+, and Cr6+, respectively. The abundance ratio of Desulfovibrio vulgaris detected by FISH to total cell counts was consistent with the obtained results of cloning and DGGE. This indicated that D. vulgaris was dominant in all analyzed samples and played a key role in heavy metal removal in R1, R2, and R3. In contrast, after 4 weeks of operation of R4, a distinct biomass loss was observed and no positive hybridized cells were detected by specific probes for the domain Bacteria, sulfate-reducing bacteria and D. vulgris. High removal efficiencies of heavy metals were achieved in R1, R2 and R3 after 12 weeks, whereas the precipitation of heavy metals in R4 was significantly decreased after 4 weeks and almost not observed after 6 weeks of operation. In addition, the anaerobic bacteria, such as Pertrimonas sulfuriphila, Clostridium sp., Citrobacter amalonaticus, and Klebsiella sp., identified from DGGE bands and clone library were hypothesized as heavy metal resistant bacteria at a loading rate of 1.5 mg l−1 d−1 of Cu2+, Ni2+, Zn2+, and Cr6+.



Similar content being viewed by others
References
Johnson DB, Hallberg DB (2005) Acid mine drainage remediation option: a review. Sci Total Environ 338:3–14
Azabou S, Mechichi T, Sayadi S (2007) Zinc precipitation by heavy-metal tolerant sulfate-reducing bacteria enriched on phosphogypsum as a sulfate source. Miner Eng 20:173–178
Smillie RH, Hunter K, Loutit M (1981) Reduction of chromium (VI) by bacterially produced hydrogen sulfide in a marine environment. Water Res 15:1351–1354
Chang IS, Kim BH (2007) Effect of sulfate reducing activity on biological treatment of hexavalent chromium [Cr(VI)] contaminated electroplating wastewater under sulfate-rich condition. Chemosphere 68:218–226
Lovley DR, Phillips EJ (1994) Reduction of chromate by Desulfovibrio vulgaris and its c 3 cytochrome. Appl Environ Microbiol 60:726–738
Goulhen F, Gloter A, Guyot F, Bruschi M (2006) Cr(VI) detoxification by Desulfovibrio vulgaris strain Hildenborough: microbe-metal interactions studies. Appl Microbiol Biotechnol 71:892–897
Lenz M, Hullebusch EDV, Hommes G, Corvini PFX, Lens PNL (2008) Selenate removal in methanogenic and sulfate-reducing upflow anaerobic sludge bed reactors. Water Res 42:2184–2194
Sahinkaya E (2009) Biotreatment of zinc-containing wastewater in sulfidogenic CSTR: performance and artificial neutral network (ANN) modeling studies. J Hazard Mater 164:105–113
Viggi CC, Pagnanelli F, Cibati A, Uccelletti D, Palleschi C, Toro L (2010) Biotreatment and bioassessment of heavy metal removal by sulfate-reducing bacteria in fixed bed reactors. Water Res 44:151–158
Li Z, Xu J, Tang C, Wu J, Muhammad A, Wang H (2006) Application of 16S rDNA-PCR amplification and DGGE fingerprinting for detection of shift in microbial community diversity in Cu-, Zn-, and Cd-contaminated paddy soils. Chemosphere 62:1374–1380
Muyzer G, De Waal ED, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M (2002) Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl Environ Microbiol 68:5064–5081
Chang YJ, Peacock AD, Long PE, Stephen JR, McKinley JP, Macnaughton SJ, Anwar Hussain AKM, Saxton AM, White DC (2001) Diversity and characterization of sulfate-reducing bacteria in groundwater at a uranium mill tailings site. Appl Environ Microbiol 67:3149–3160
Nakagawa T, Hanada S, Maruyama A, Marumo K, Urabe T, Fukui M (2002) Distribution and diversity of thermophilic sulfate-reducing bacteria within a Cu-Pb-Zn mine (Toyoha, Japan). Microbiol Ecol 41:199–209
Kaksonen AH, Plumb JJ, Franzmann PD, Puhakka JA (2004) Simple organic electron donors support diverse sulfate-reducing communities in fluidized-bed reactors treating acidic metal-and sulfate-containing wastewater. Microbiol Ecol 47:279–289
Poulson SR, Colberg PJS, Drever JI (1997) Toxicity of heavy metal (Ni, Zn) to Desulfovibrio desulfuricans. J Geomicrobiol 14:41–49
Cabrera G, Pérez JM, Gómez A, Canterro D (2006) Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J Hazard Mater A135:40–46
Postgate JR (1984) The sulfate-reducing bacteria, 2nd edn. Cambridge University Press, Cambridge
Kieu TQH, Müller E, Horn H (2011) Heavy metal removal in anaerobic semi-continuous stirred tank reactors by a consortium of sulfate-reducing bacteria. Water Res 45(13):3863–3870
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic, John Wiley & Sons, Chichester, United Kingdom, pp 115–175
Nielsen AT, Liu WT, Filipe C, Grady L, Molin S, Stahl DA (1999) Identification of a novel group of bacteria in sludge from a deteriorated biological phosphorus removal reactor. Appl Environ Microbiol 65:1251–1258
Geets J, Borremans B, Diels L, Springael D, Vangronsveld J, Van der Lelie D, Vanbroekhoven K (2006) DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J Microbiol Methods 66:194–205
Wagner M, Roger AJ, Flax JL, Brusseau GA, Stahl DA (1998) Phylogeny of dissimilatory sulfite reductase supports an early origin of sulfate respiration. J Bacteriol 180:2975–2982
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresiss (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141
Don RH, Wainwright BJ, Baker K, Mattick JS (1991) “Touchdown” PCR to circumvent spurious priming during gene amplification. Nucl Acids Res 19:4008
Altschul SF, Madden TL, Schäfer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 27:3389–33402
Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussman R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenkej M, Ludwig T, Bode A, Schleifer KH (2004) ARB: a software environment for sequence data. Nucl Acids Res 32:1363–1371
Amann R, Stromley J, Devereux R, Key R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Daims H, Brühl A, Amann R, Schleifer KH, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Wallner G, Amann R, Beisker W (1993) Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136–143
Stahl DA and Amann R (1991) Development and application of nucleic acid probes. In: Stackebrandt EM, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom, pp 205–248
Deveruex RM, Kane D, Winfrey J, Stahl DA (1992) Genus-and group-specific hybridization probes for determinative and envioronmental studies of sulfate-reducing bacteria. Syst Appl Microbiol 15:601–609
Manz W, Eisenbrecher M, Neu TR, Szewzyk U (1998) Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol 25:43–61
Brosius J, Dull TJ, Sleeter DD, Noller HF (1981) Gene organization and primary structure of a ribosomal RNA operon from E. coli. J Mol Biol 148:107–127
Azabou S, Mechichi T, Sayadi S (2007) Zinc precipitation by heavy-metal tolerant sulfate-reducing bacteria enriched on phosphogypsum as a sulfate source. Mineral Eng 20:173–178
Hao OJ, Huang L, Chen JM, Bugass RL (1994) Effects of metal additions on sulfate reduction activity in wastewater. Toxicol Environ Chem 46(4):197–212
Utgikar VP, Chaudhary N, Koeniger A, Tabax HH, Heines JR, Govind R (2004) Toxicity of metals and metal mixtures analysis of concentration and time dependence for zinc and copper. Water Res 38:3651–3658
Dar SA, Bijmans MF, Dinkla IJ, Geurkink B, Lens PN, Dopson M (2009) Population dynamics of a single-stage sulfidogenic bioreactor treating synthetic zinc-containing waste streams. Microbiol Ecol 58:529–537
Pan J, Yu L (2011) Effects of Cd or/and Pb on soil enzyme activities and microbial community structure. Ecol Eng 37:1889–1894
Michel C, Brugna M, Aubert C, Bernadac A, Bruschi M (2001) Enzymatic reduction of chromate: comparative studies using sulfate-reducing bacteria. Appl Microbiol Biotechnol 55:95–100
Acknowledgments
The authors acknowledge financial support provided by Federal Ministry for Education and Research (BMBF/Germany) and Institute of Water Quality Control, Faculty of Civil Engineering and Geodesy, Technische Universität München, Germany.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kieu, H.T.Q., Horn, H. & Müller, E. The effect of heavy metals on microbial community structure of a sulfidogenic consortium in anaerobic semi-continuous stirred tank reactors. Bioprocess Biosyst Eng 37, 451–460 (2014). https://doi.org/10.1007/s00449-013-1012-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-1012-5