Abstract
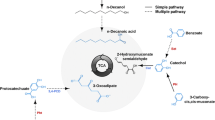
Streptomyces avermitilis is a well known organism producing avermectin antibiotics, and has been utilized as an industrial host for oxidation bioconversion processes. Recently, gene screening strategies related to bioconversions have received much focus, as attempts are made to optimize oxidation and biodegradation pathways to maximize yield and productivity. Here, we have demonstrated the oxidative metabolisms of three molecules, daidzein, p-coumaric acid and mevastatin, where S. avermitilis converted each substrate to 3′,4′,7-trihydroxyisoflavone, caffeic acid and hydroxyl-mevastatin to yield 9.3, 32.5 and 15.0 %, respectively. Microarray technology was exploited to investigate genome-wide analysis of gene expression changes, which were induced upon the addition of each substrate. Cytochrome P450 hydroxylases (pteC, cyp28 and olmB), diooxygenases (xylE, cdo1 and putatives) and LuxAB-like oxygenase were identified. One of them, cyp28, was indeed a gene encoding P450 hydroxylase responsible for the oxidative reaction of daidzein. Furthermore, possible electron transfer chain (fdrC → pteE → pteC) supporting cytochrome P450 dependent hydroxylation of daidzein has been suggested based on the interpretation of expression profiles. The result provided a potential application of transcriptomic study on uncovering enzymes involved in oxidative bioconversions of S. avermitilis.





Similar content being viewed by others
References
Paradkar A, Trefzer A, Chakraburtty R, Stassi D (2003) Streptomyces genetics: a genomic perspective. Crit Rev Biotechnol 23:1–27
Chen M, Wang G, Dai S, Xie L, Li X (2009) Polyketide antibiotics produced by polyketide synthase in streptomyces–a review. Wei Sheng Wu Xue Bao = Acta Microbiol Sinica 49:1555–1563
Horvath I, Lovrekovich I, Varga JM (1964) Antibiotics produced by Streptomyces. 3. A new antibiotic K 178; biological studies. Z Allg Mikrobiol 61:236–241
Kim YS, Kim HM, Chang C, Hwang IC, Oh H, Ahn JS, Kim KD, Hwang BK, Kim BS (2007) Biological evaluation of neopeptins isolated from a Streptomyces strain. Pest Manag Sci 63:1208–1214
Rassi H, Ghasemi SF (2008) Production of novel chemotherapeutic drugs by Streptomycetes. Lik Sprava 7–8:40–47
Zhan J (2009) Biosynthesis of bacterial aromatic polyketides. Curr Top Med Chem 9:1610–1958
Ikeda H, Kotaki H, Omura S (1987) Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J Bacteriol 169:5615–5621
MacNeil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, MacNeil T (1992) Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Ikeda H, Omura S (1995) Control of avermectin biosynthesis in Streptomyces avermitilis for the selective production of a useful component. J Antibiot (Tokyo) 48:549–562
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci USA 98:12215–12220
Quaderer R, Omura S, Ikeda H, Cane DE (2006) Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlI, a cytochrome P450 of Streptomyces avermitilis. J Am Chem Soc 128:13036–13037
Lamb DC, Ikeda H, Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL (2003) Cytochrome p450 complement (CYPome) of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem Biophys Res Commun 307:610–619
Pandey BP, Roh C, Choi KY, Lee N, Kim EJ, Ko S, Kim T, Yun H, Kim BG (2010) Regioselective hydroxylation of daidzein using P450 (CYP105D7) from Streptomyces avermitilis MA4680. Biotechnol Bioeng 105:697–704
Roh C, Seo SH, Choi KY, Cha M, Pandey BP, Kim JH, Park JS, Kim DH, Chang IS, Kim BG (2009) Regioselective hydroxylation of isoflavones by Streptomyces avermitilis MA-4680. J Biosci Bioeng 108:41–46
Bugg TD, Ahmad M, Hardiman EM, Rahmanpour R (2011) Pathways for degradation of lignin in bacteria and fungi. Nat Prod Rep 28:1883–1896
Crawford DL, Pometto AL 3rd, Crawford RL (1984) Production of useful modified lignin polymers by bioconversion of lignocellulose with Streptomyces. Biotechnol Adv 2:217–232
Davis JR, Sello JK (2010) Regulation of genes in Streptomyces bacteria required for catabolism of lignin-derived aromatic compounds. Appl Microbiol Biotechnol 86:921–929
Iwagami SG, Yang K, Davies J (2000) Characterization of the protocatechuic acid catabolic gene cluster from Streptomyces sp. strain 2065. Appl Environ Microbiol 66:1499–1508
Stanier RY (1950) The bacterial oxidation of aromatic compounds. IV. Studies on the mechanism of enzymatic degradation of protocatechuic acid. J Bacteriol 59:527–532
Rivas FJ (2006) Polycyclic aromatic hydrocarbons sorbed on soils: a short review of chemical oxidation based treatments. J Hazard Mater 138:234–251
Artham T, Doble M (2008) Biodegradation of aliphatic and aromatic polycarbonates. Macromol Biosci 8:14–24
Murray M (1999) Mechanisms and significance of inhibitory drug interactions involving cytochrome P450 enzymes. Int J Mol Med 3:227–238
Chen W, He F, Zhang X, Chen Z, Wen Y, Li J (2010) Chromosomal instability in Streptomyces avermitilis: major deletion in the central region and stable circularized chromosome. BMC Microbiol 10:198
Yun J, Ryu S (2005) Screening for novel enzymes from metagenome and SIGEX, as a way to improve it. Microb Cell Fact 4:8
Chigu NL, Hirosue S, Nakamura C, Teramoto H, Ichinose H, Wariishi H (2010) Cytochrome P450 monooxygenases involved in anthracene metabolism by the white-rot basidiomycete Phanerochaete chrysosporium. Appl Microbiol Biotechnol 87:1907–1916
Syed K, Doddapaneni H, Subramanian V, Lam YW, Yadav JS (2010) Genome-to-function characterization of novel fungal P450 monooxygenases oxidizing polycyclic aromatic hydrocarbons (PAHs). Biochem Biophys Res Commun 399:492–497
Watanabe I, Nara F, Serizawa N (1995) Cloning, characterization and expression of the gene encoding cytochrome P-450sca-2 from Streptomyces carbophilus involved in production of pravastatin, a specific HMG-CoA reductase inhibitor. Gene 163:81–85
Shepherd MD, Kharel MK, Bosserman MA, Rohr J (2010) In: Coico R, Kowalik T, Quarles J, Stevenson B, Taylor R (eds) Current protocols in microbiology. J. Wiley & Sons, NJ
Kulling SE, Honig DM, Metzler M (2001) Oxidative metabolism of the soy isoflavones daidzein and genistein in humans in vitro and in vivo. J Agric Food Chem 49:3024–3033
Choi KY, Jung E, Jung DH, An BR, Pandey BP, Yun H, Sung C, Park HY, Kim BG (2012) Engineering of daidzein 3′-hydroxylase P450 enzyme into catalytically self-sufficient cytochrome P450. Microb Cell Fact 11:81
Kumar RP, Ravindranath SD, Vaidyanathan CS, Rao NA (1972) Mechanism of hydroxylation of aromatic compounds. II. Evidence for the involvement of superoxide anions in enzymatic hydroxylations. Biochem Biophys Res Commun 49:1422–1426
Mitoma C, Posner HS, Reitz HC, Udenfriend S (1956) Enzymatic hydroxylation of aromatic compounds. Arch Biochem Biophys 61:431–441
Ullrich R, Hofrichter M (2007) Enzymatic hydroxylation of aromatic compounds. Cell Mol Life Sci 64:271–293
Choi KY, Jung E, Jung DH, Pandey BP, Yun H, Park HY, Kazlauskas RJ, Kim BG (2012) Cloning, expression and characterization of CYP102D1, a self-sufficient P450 monooxygenase from Streptomyces avermitilis. FEBS J 279:1650–1662
Sevrioukova IF, Li H, Zhang H, Peterson JA, Poulos TL (1999) Structure of a cytochrome P450-redox partner electron-transfer complex. Proc Natl Acad Sci USA 96:1863–1868
Acknowledgments
The authors wish to acknowledge the financial support of the Seoul R&BD Program (KU080657) and WCU (World Class University) program through the National Research Foundation of Korea (NRF) grant funded by MEST (R322012000102130) and also through Institute of Bioengineering, Seoul National University, Seoul, Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
H.-J. Kim and K.-Y. Choi contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, HJ., Choi, KY., Jung, DH. et al. Transcriptomic study for screening genes involved in the oxidative bioconversions of Streptomyces avermitilis . Bioprocess Biosyst Eng 36, 1621–1630 (2013). https://doi.org/10.1007/s00449-013-0935-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00449-013-0935-1