Abstract
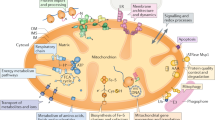
Mitochondria play a key role in several metabolic and cell biological pathways and have attracted increasing attention due to their implication in life-span, ageing and human diseases. Mitochondrial proteases have a special role in these multiple biological functions, as they are involved in the regulation of various processes, e.g., mitochondrial protein biogenesis and quality control, mitochondrial dynamics, mitophagy and programmed cell death. The mitochondrial presequence processing machinery serves the particular purpose of maturing the majority of incoming precursor proteins by presequence cleavage, to ensure a stable mature protein by trimming of intermediate N-termini and to remove free toxic targeting peptides.




Similar content being viewed by others
References
Alikhani N, Berglund AK, Engmann T, Spånning E, Vögtle FN, Pavlov P, Meisinger C, Langer T, Glaser E (2011) Targeting capacity and conservation of PreP homologues localization in mitochondria of different species. J Mol Biol 410:400–410
Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, Langer T (2014) The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol 204:919–929
Arlt H, Tauer R, Feldmann H, Neupert W, Langer T (1996) The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875–885
Bhushan S, Ståhl A, Nilsson S, Lefebvre B, Seki M, Roth C, McWilliam D, Wright SJ, Liberles DA, Shinozaki K, Bruce BD, Boutry M, Glaser E (2005) Catalysis, subcellular localization, expression and evolution of the targeting peptides degrading protease, AtPreP2. Plant Cell Physiol 46:985–996
Bonn F, Tatsuta T, Petrungaro C, Riemer J, Langer T (2011) Presequence-dependent folding ensures MrpL32 processing by the m-AAA protease in mitochondria. EMBO J 30:2545–2556
Braun HP, Emmermann M, Kruft V, Schmitz UK (1992) The general mitochondrial processing peptidase from potato is an integral part of cytochrome c reductase of the respiratory chain. EMBO J 11:3219–3227
Brunetti D, Torsvik J, Dallabona C, Teixeira P, Sztromwasser P, Fernandez-Vizarra E, Cerutti R, Reyes A, Preziuso C, D’Amati G, Baruffini E, Goffrini P, Viscomi C, Ferrero I, Boman H, Telstad W, Johansson S, Glaser E, Knappskog PM, Zeviani M, Bindoff LA (2015) Defective PITRM1 mitochondrial peptidase is associated with Aβ amyloidotic neurodegeneration. EMBO Mol Med 8:176–190
Büchler M, Tisljar U, Wolf DH (1994) Proteinase yscD (oligopeptidase yscD). Structure, function and relationship of the yeast enzyme with mammalian thimet oligopeptidase (metalloendopeptidase, EP 24.15). Eur J Biochem 219:627–639
Burkhart JM, Taskin AA, Zahedi RP, Vögtle FN (2015) Quantitative profiling for substrates of the mitochondrial presequence processing protease reveals a set of nonsubstrate proteins Increased upon proteotoxic stress. J Proteome Res 14:4550–4563
Carrie C, Venne AS, Zahedi RP, Soll J (2015) Identification of cleavage sites and substrate proteins for two mitochondrial intermediate peptidases in Arabidopsis thaliana. J Exp Bot 66:2691–2708
Choquet K, Zurita-Rendón O, La Piana R, Yang S, Dicaire MJ, Care4Rare Consortium, Boycott KM, Majewski J, Shoubridge EA, Brais B, Tétreault M (2016) Autosomal recessive cerebellar ataxia caused by a homozygous mutation in PMPCA. Brain 139(Pt 3):e19. doi:10.1093/brain/awv362
Dvoráková-Holá K, Matusková A, Kubala M, Otyepka M, Kucera T, Vecer J, Herman P, Parkhomenko N, Kutejova E, Janata J (2010) Glycine-rich loop of mitochondrial processing peptidase alpha-subunit is responsible for substrate recognition by a mechanism analogous to mitochondrial receptor Tom20. J Mol Biol 396:1197–1210
Erşahin C, Szpaderska AM, Orawski AT, Simmons WH (2005) Aminopeptidase P isozyme expression in human tissues and peripheral blood mononuclear cell fractions. Arch Biochem Biophys 435:303–310
Esser K, Pratje E, Michaelis G (1996) SOM 1, a small new gene required for mitochondrial inner membrane peptidase function in Saccharomyces cerevisiae. Mol Gen Genet 252:437–445
Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E (2002) A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol 323:835–843
Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Eneqvist T, Tjernberg L, Ankarcrona M, Glaser E (2006) Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem 281:29096–29104
Fang D, Wang Y, Zhang Z, Du H, Yan S, Sun Q, Zhong C, Wu L, Vangavaragu JR, Yan S, Hu G, Guo L, Rabinowitz M, Glaser E, Arancio O, Sosunov AA, McKhann GM, Chen JX, Yan SS (2015) Increased neuronal PreP activity reduces Aβ accumulation, attenuates neuroinflammation and improves mitochondrial and synaptic function in Alzheimer disease’s mouse model. Hum Mol Genet 24:5198–5210
Freeman M (2014) The rhomboid-like superfamily: molecular mechanisms and biological roles. Annu Rev Cell Dev Biol 30:235–254
Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K (2015) MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics 14:1113–1126
Gakh O, Cavadini P, Isaya G (2002) Mitochondrial processing peptidases. Biochim Biophys Acta 1592:63–77
Gerdes F, Tatsuta T, Langer T (2012) Mitochondrial AAA proteases--towards a molecular understanding of membrane-bound proteolytic machines. Biochim Biophys Acta 1823:49–55
Gray MW (2015) Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci U S A 112:10133–10138
Habib SJ, Neupert W, Rapaport D (2007) Analysis and prediction of mitochondrial targeting signals. Methods Cell Biol 80:761–781
Harbauer AB, Zahedi RP, Sickmann A, Pfanner N, Meisinger C (2014) The protein import machinery of mitochondria-a regulatory hub in metabolism, stress, and disease. Cell Metab 19:357–372
Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl FU, Neupert W (1988) Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell 53:795–806
Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS (2003) Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem 278:27781–27788
Huang S, Taylor NL, Whelan J, Millar AH (2009) Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol 150:1272–1285
Huang S, Nelson CJ, Li L, Taylor NL, Ströher E, Peteriet J, Millar AH (2015) Intermediate cleavage peptidase55 Modifies Enzyme Amino Termini and Alters Protein Stability in Arabidopsis Mitochondria. Plant Physiol 168:415–27
Ieva R, Heißwolf AK, Gebert M, Vögtle FN, Wollweber F, Mehnert CS, Oeljeklaus S, Warscheid B, Meisinger C, van der Laan M, Pfanner N (2013) Mitochondrial inner membrane protease promotes assembly of presequence translocase by removing a carboxy-terminal targeting sequence. Nat Commun 4:2853
Isaya G, Kalousek F, Fenton WA, Rosenberg LE (1991) Cleavage of precursors by the mitochondrial processing peptidase requires a compatible mature protein or an intermediate octapeptide. J Cell Biol 113:65–76
Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ (2010) Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol 191:933–942
Jobling RK, Assoum M, Gakh O, Blaser S, Raiman JA, Mignot C, Roze E, Dürr A, Brice A, Lévy N, Prasad C, Paton T, Paterson AD, Roslin NM, Marshall CR, Desvignes JP, Roëckel-Trevisiol N, Scherer SW, Rouleau GA, Mégarbané A, Isaya G, Delague V, Yoon G (2015) PMPCA mutations cause abnormal mitochondrial protein processing in patients with non-progressive cerebellar ataxia. Brain 138:1505–1517
Johnson KA, Bhushan S, Ståhl A, Hallberg BM, Frohn A, Glaser E, Eneqvist T (2006) The closed structure of presequence protease PreP forms a unique 10,000 Angstroms3 chamber for proteolysis. EMBO J 25:1977–1986
Joshi M, Anselm I, Shi J, Bale TA, Towne M, Schmitz-Abe K, Crowley L, Giani FC, Kazerounian S, Markianos K, Lidov HG, Folkerth R, Sankaran VG, Agrawal PB (2016) Mutations in the substrate binding glycine-rich loop of the mitochondrial processing peptidase-α protein (PMPCA) cause a severe mitochondrial disease. Cold Spring Harb Mol Case Stud 2:a000786. doi:10.1101/mcs.a000786
Kalousek F, Hendrick JP, Rosenberg LE (1988) Two mitochondrial matrix proteases act sequentially in the processing of mammalian matrix enzymes. Proc Natl Acad Sci U S A 85:7536–7540
Kambacheld M, Augustin S, Tatsuta T, Müller S, Langer T (2005) Role of the novel metallopeptidase Mop112 and saccharolysin for the complete degradation of proteins residing in different subcompartments of mitochondria. J Biol Chem 280:20132–20139
King JV, Liang WG, Scherpelz KP, Schilling AB, Meredith SC, Tang WJ (2014) Molecular basis of substrate recognition and degradation by human presequence protease. Structure 22:996–1007
Kmiec B, Teixeira PF, Berntsson RP, Murcha MW, Branca RM, Radomiljac JD, Regberg J, Svensson LM, Bakali A, Langel U, Lehtiö J, Whelan J, Stenmark P, Glaser E (2013) Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc Natl Acad Sci U S A 110:3761–3769
Kmiec B, Teixeira PF, Glaser E (2014) Shredding the signal: targeting peptide degradation in mitochondria and chloroplasts. Trends Plant Sci 19:771–778
Kučera T, Otyepka M, Matušková A, Samad A, Kutejová E, Janata J (2013) A computational study of the glycine-rich loop of mitochondrial processing peptidase. PLoS ONE 8:e74518
Lee CM, Sedman J, Neupert W, Stuart RA (1999) The DNA helicase, Hmi1p, is transported into mitochondria by a C-terminal cleavable targeting signal. J Biol Chem 274:20937–20942
Leissring MA (2014) Aβ degradation-the inside story. Front Aging Neurosci 6:229
Lightowlers RN, Taylor RW, Turnbull DM (2015) Mutations causing mitochondrial disease: What is new and what challenges remain? Science 349:1494–1499
Lionaki E, Tavernarakis N (2013) Oxidative stress and mitochondrial protein quality control in aging. J Proteomics 92:181–194
Luo W, Fang H, Green N (2006) Substrate specificity of inner membrane peptidase in yeast mitochondria. Mol Genet Genomics 275:431–436
Martelli A, Puccio H (2014) Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front Pharmacol 5:130
McQuibban GA, Saurya S, Freeman M (2003) Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature 423:537–541
Meisinger C, Sickmann A, Pfanner N (2008) The mitochondrial proteome: from inventory to function. Cell 134:22–24
Moberg P, Ståhl A, Bhushan S, Wright SJ, Eriksson A, Bruce BD, Glaser E (2003) Characterization of a novel zinc metalloprotease involved in degrading targeting peptides in mitochondria and chloroplasts. Plant J 36:616–628
Mossmann D, Meisinger C, Vögtle FN (2012) Processing of mitochondrial presequences. Biochim Biophys Acta 1819:1098–1106
Mossmann D, Vögtle FN, Taskin AA, Teixeira PF, Ring J, Burkhart JM, Burger N, Pinho CM, Tadic J, Loreth D, Graff C, Metzger F, Sickmann A, Kretz O, Wiedemann N, Zahedi RP, Madeo F, Glaser E, Meisinger C (2014) Amyloid-β peptide induces mitochondrial dysfunction by inhibition of preprotein maturation. Cell Metab 20:662–669
Neupert W (2015) A perspective on transport of proteins into mitochondria: a myriad of open questions. J Mol Biol 427:1135–1158
Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T (2005) The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell 123:277–289
Nunnari J, Fox TD, Walter P (1993) A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science 262:1997–2004
Osman C, Wilmes C, Tatsuta T, Langer T (2007) Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase. Mol Biol Cell 18:627–635
O’Toole JF, Liu Y, Davis EE, Westlake CJ, Attanasio M, Otto EA, Seelow D, Nurnberg G, Becker C, Nuutinen M, Kärppä M, Ignatius J, Uusimaa J, Pakanen S, Jaakkola E, van den Heuvel LP, Fehrenbach H, Wiggins R, Goyal M, Zhou W, Wolf MT, Wise E, Helou J, Allen SJ, Murga-Zamalloa CA, Ashraf S, Chaki M, Heeringa S, Chernin G, Hoskins BE, Chaib H, Gleeson J, Kusakabe T, Suzuki T, Isaac RE, Quarmby LM, Tennant B, Fujioka H, Tuominen H, Hassinen I, Lohi H, van Houten JL, Rotig A, Sayer JA, Rolinski B, Freisinger P, Madhavan SM, Herzer M, Madignier F, Prokisch H, Nurnberg P, Jackson PK, Khanna H, Katsanis N, Hildebrandt F (2010) Individuals with mutations in XPNPEP3, which encodes a mitochondrial protein, develop a nephronophthisis-like nephropathy. J Clin Invest 120:791–802
Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123
Pinho CM, Teixeira PF, Glaser E (2014) Mitochondrial import and degradation of amyloid-β peptide. Biochim Biophys Acta 1837:1069–1074
Potting C, Wilmes C, Engmann T, Osman C, Langer T (2010) Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J 29:2888–2898
Quirós PM, Langer T, López-Otín C (2015) New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol 16:345–359
Reinders J, Zahedi RP, Pfanner N, Meisinger C, Sickmann A (2006) Toward the complete yeast mitochondrial proteome: multidimensional separation techniques for mitochondrial proteomics. J Proteome Res 5:1543–1554
Rhein V, Song X, Wiesner A, Ittner LM, Baysang G, Meier F, Ozmen L, Bluethmann H, Dröse S, Brandt U, Savaskan E, Czech C, Götz J, Eckert A (2009) Amyloid-beta and tau synergistically impair the oxidative phosphorylation system in triple transgenic Alzheimer’s disease mice. Proc Natl Acad Sci U S A 106:20057–20062
Riemer J, Fischer M, Herrmann JM (2011) Oxidation-driven protein import into mitochondria: Insights and blind spots. Biochim Biophys Acta 1808:981–989
Rugarli EI, Langer T (2012) Mitochondrial quality control: a matter of life and death for neurons. EMBO J 31:1336–1349
Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G (1991) Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J 10:247–254
Sokol AM, Sztolsztener ME, Wasilewski M, Heinz E, Chacinska A (2014) Mitochondrial protein translocases for survival and wellbeing. FEBS Lett 588:2484–2495
Stahl A, Moberg P, Ytterberg J, Panfilov O, Brockenhuus Von Lowenhielm H, Nilsson F, Glaser E (2002) Isolation and identification of a novel mitochondrial metalloprotease (PreP) that degrades targeting presequences in plants. J Biol Chem 277:41931–4139
Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T (2007) m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J 26:325–335
Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J (2001) Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure 9:615–625
Teixeira PF, Glaser E (2013) Processing peptidases in mitochondria and chloroplasts. Biochim Biophys Acta 1833:360–370
Vangavaragu JR, Valasani KR, Gan X, Yan SS (2014) Identification of human presequence protease (hPreP) agonists for the treatment of Alzheimer’s disease. Eur J Med Chem 76:506–516
Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20:1298–1345
Venne AS, Vögtle FN, Meisinger C, Sickmann A, Zahedi RP (2013) Novel highly sensitive, specific, and straightforward strategy for comprehensive N-terminal proteomics reveals unknown substrates of the mitochondrial peptidase Icp55. J Proteome Res 12:3823–3830
Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139:428–439
Vögtle FN, Prinz C, Kellermann J, Lottspeich F, Pfanner N, Meisinger C (2011) Mitochondrial protein turnover: role of the precursor intermediate peptidase Oct1 in protein stabilization. Mol Biol Cell 22:2135–2143
Vögtle FN, Burkhart JM, Rao S, Gerbeth C, Hinrichs J, Martinou JC, Chacinska A, Sickmann A, Zahedi RP, Meisinger C (2012) Intermembrane space proteome of yeast mitochondria. Mol Cell Proteomics 11:1840–1852
Weckbecker D, Longen S, Riemer J, Herrmann JM (2012) Atp23 biogenesis reveals a chaperone-like folding activity of Mia40 in the IMS of mitochondria. EMBO J 31:4348–4358
Yao J, Irwin RW, Zhao L, Nilsen J, Hamilton RT, Brinton RD (2009) Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A 106:14670–14675
Yang M, Jensen RE, Yaffe MP, Oppliger W, Schatz G (1988) Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J 7:3857–3862
Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T (2001) Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science 291:2135–2138
Zeng X, Kucharczyk R, di Rago JP, Tzagoloff A (2007) The leader peptide of yeast Atp6p is required for efficient interaction with the Atp9p ring of the mitochondrial ATPase. J Biol Chem 282:36167–36176
Acknowledgment
F.N.V. is supported by the Emmy-Noether-Program of the DFG (Deutsche Forschungsgemeinschaft).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Poveda-Huertes, D., Mulica, P. & Vögtle, F.N. The versatility of the mitochondrial presequence processing machinery: cleavage, quality control and turnover. Cell Tissue Res 367, 73–81 (2017). https://doi.org/10.1007/s00441-016-2492-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-016-2492-9