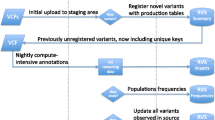
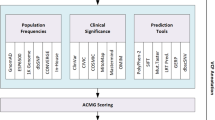
Abstract
With the increasing importance of genomic data in understanding genetic diseases, there is an essential need for efficient and user-friendly tools that simplify variant analysis. Although multiple tools exist, many present barriers such as steep learning curves, limited reference genome compatibility, or costs. We developed VARista, a free web-based tool, to address these challenges and provide a streamlined solution for researchers, particularly those focusing on rare monogenic diseases. VARista offers a user-centric interface that eliminates much of the technical complexity typically associated with variant analysis. The tool directly supports VCF files generated using reference genomes hg19, hg38, and the emerging T2T, with seamless remapping capabilities between them. Features such as gene summaries and links, tissue and cell-specific gene expression data for both adults and fetuses, as well as automated PCR design and integration with tools such as SpliceAI and AlphaMissense, enable users to focus on the biology and the case itself. As we demonstrate, VARista proved effective in narrowing down potential disease-causing variants, prioritizing them effectively, and providing meaningful biological context, facilitating rapid decision-making. VARista stands out as a freely available and comprehensive tool that consolidates various aspects of variant analysis into a single platform that embraces the forefront of genomic advancements. Its design inherently supports a shift in focus from technicalities to critical thinking, thereby promoting better-informed decisions in genetic disease research. Given its unique capabilities and user-centric design, VARista has the potential to become an essential asset for the genomic research community. https://VARista.link


Similar content being viewed by others
Availability of data and materials
VARista is available at: https://VARista.link. The code for VARista, including all the databases it utilizes is hosted at Zendo: https://zenodo.org/record/8384364. The code for ViCiFier, an Illumina sequencing raw data preprocessing pipeline from FASTQ to VCF, is hosted at https://github.com/Noam-Hadar/ViCiFier
Abbreviations
- VCF:
-
Variant call format
- PCR:
-
Polymerase chain reaction
- T2T:
-
Telomere-to-telomere
- SNV:
-
Single nucleotide variation
- INDEL:
-
Insertion/deletion
- SV:
-
Structural variant
- HPO:
-
Human phenotype ontology
- HGVS:
-
Human Genome Variation Society
References
Aganezov S et al (2022) A complete reference genome improves analysis of human genetic variation. Science (1979). https://doi.org/10.1126/science.abl3533
Bamshad MJ, Nickerson DA, Chong JX (2019) Mendelian gene discovery: fast and furious with no end in sight. Am J Hum Genet 105:448–455
Bateman A et al (2023) UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res 51:D523–D531
Blake JA et al (2021) Mouse genome database (MGD): knowledgebase for mouse-human comparative biology. Nucleic Acids Res 49:D981–D987
Cao J et al (2020) A human cell atlas of fetal gene expression. Science. https://doi.org/10.1126/science.aba7721
Cheng J et al (2023) Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science (1979). https://doi.org/10.1126/SCIENCE.ADG7492
Cingolani P et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80
Davieson CD, Joyce KE, Sharma L, Shovlin CL (2021) DNA variant classification–reconsidering “allele rarity” and “phenotype” criteria in ACMG/AMP guidelines. Eur J Med Genet 64:104312
den Dunnen JT et al (2016) HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat 37:564–569
Fakhro KA et al (2016) The Qatar genome: a population-specific tool for precision medicine in the Middle East. Hum Genome Var. https://doi.org/10.1038/hgv.2016.16
Fishilevich S et al (2017) GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017:1–17
Garcia FAdO, Andrade ESd, Palmero EI (2022) Insights on variant analysis in silico tools for pathogenicity prediction. Front Genet 13:1010327
Gombosh M et al (2023) De-novo “germline second hit” loss-of-heterozygosity RBP3 deletion mutation causing recessive high myopia. Clin Genet. https://doi.org/10.1111/CGE.14384
Groza T et al (2022) The International Mouse Phenotyping Consortium: comprehensive knockout phenotyping underpinning the study of human disease. Nucleic Acids Res. https://doi.org/10.1093/NAR/GKAC972
Gudmundsson S et al (2021) Variant interpretation using population databases: lessons from gnomAD. Hum Mutat. https://doi.org/10.1002/HUMU.24309
Hadar N, Weintraub G, Gudes E, Dolev S, Birk OS (2023a) GeniePool: genomic database with corresponding annotated samples based on a cloud data lake architecture. Database (Oxford). https://doi.org/10.1093/database/baad043
Hadar N et al (2023b) X-linked C1GALT1C1 mutation causes atypical hemolytic uremic syndrome. Eur J Hum Genet 2023:1–7. https://doi.org/10.1038/s41431-022-01278-5
Hadar N et al (2024) Heterozygous THBS2 pathogenic variant causes Ehlers-Danlos syndrome with prominent vascular features in humans and mice. Eur J Hum Genet. https://doi.org/10.1038/S41431-024-01559-1
Hoyt SJ et al (2022) From telomere to telomere: the transcriptional and epigenetic state of human repeat elements. Science (1979) 376:eabk3112
Jain M, Olsen HE, Paten B, Akeson M (2016) The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 17:1–11
Kent WJ et al (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kishore Jaganathan A et al (2018) Predicting splicing from primary sequence with deep learning in brief a deep neural network precisely models mRNA splicing from a genomic sequence and accurately predicts noncoding cryptic splice mutations in patients with rare genetic diseases. Predicting splicing from primary sequence with deep learning. Cell 176:535–54824
Köhler S et al (2021) The human phenotype ontology in 2021. Nucleic Acids Res 49:D1207–D1217
Kopanos C et al (2019) VarSome: the human genomic variant search engine. Bioinformatics 35:1978
Kristal E et al (2022) Hyper IgM in tricho-hepato-enteric syndrome due to TTC37 mutation. Immunol Res 70:775–780
Landrum MJ et al (2018) ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res 46:D1062–D1067
Lee J et al (2022) A database of 5305 healthy Korean individuals reveals genetic and clinical implications for an East Asian population. Exp Mol Med 54:1862–1871
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49:D458–D460
Lonsdale J et al (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585
McKusick VA (2007) Mendelian inheritance in man and its online version. OMIM Am J Hum Genet 80:588–604. https://doi.org/10.1086/514346
O’Leary NA et al (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44:D733–D745
Pais LS et al (2022) seqr: a web-based analysis and collaboration tool for rare disease genomics. Hum Mutat 43:698–707
Phan L, Jin Y, Zhang H, Qiang W, Shekhtman E, Shao D, Revoe D, Villamarin R, Ivanchenko E, Kimura M, Wang ZY, Hao L, Sharopova N, Bihan M, Sturcke A, Lee M, Popova N, Wu W, Bastiani C, Ward M, Holmes JB, Lyoshin V, Kaur K, Moyer E, Feolo M, Kattman BL (2020) 'ALFA: Allele Frequency Aggregator.' National Center for Biotechnology Information, U.S. National Library of Medicine, https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/
Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A (2010) Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res 20:110
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteom Bioinform 13:278–289
Richards S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–424
Robinson JT et al (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26. https://doi.org/10.1038/nbt.1754
Safran A et al (2023) Hyperinsulinism/hyperammonemia syndrome caused by biallelic SLC25A36 mutation. J Inherit Metab Dis 46:744–755
Stelzer G et al (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform 2016:1.30.1-1.30.33
The cost of sequencing a human genome. (2021). https://www.genome.gov/about-genomics/fact-sheets/Sequencing-Human-Genome-cost.
Uhlen M et al (2015) Tissue-based map of the human proteome. Science 1979(347):1260419–1260419
Untergasser A et al (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res. https://doi.org/10.1093/nar/gks596
Van der Auwera GA et al (2013) From fastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.bi1110s43
Weber JL, Myers EW (1997) Human whole-genome shotgun sequencing. Genome Res 7:401–409
Ye J et al (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134
Ziv M, Gruber G, Sharon M, Vinogradov E, Yeger-Lotem E (2022) The TissueNet vol 3 database: protein-protein interactions in adult and embryonic human tissue contexts. J Mol Biol 434:167532
Acknowledgements
The authors wish to thank Prof. Eitan Rubin and Grisha Weintraub for numerous serendipitous chats which assisted in the making of this project.
Funding
The study was supported by the Morris Kahn Family Foundation, by Israel Science Foundation grant 2463/23 and by the National Knowledge Center for Rare/Orphan Diseases of the Israel Ministry of Science, Technology and Space, Ben-Gurion University of the Negev, Beer-Sheva, Israel.
Author information
Authors and Affiliations
Contributions
NH idealized, planned and developed the tool. NH and KO maintain the tool. All authors provided continuous feedback during the development of the tool. OSB supervised the project.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hadar, N., Dolgin, V., Oustinov, K. et al. VARista: a free web platform for streamlined whole-genome variant analysis across T2T, hg38, and hg19. Hum. Genet. (2024). https://doi.org/10.1007/s00439-024-02671-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00439-024-02671-4