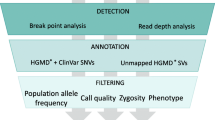
Abstract
Next-generation sequencing (NGS) is an incredibly useful tool for genetic disease diagnosis. However, the most commonly used bioinformatics methods for analyzing sequence reads insufficiently discriminate genomic regions with extensive sequence identity, such as gene families and pseudogenes, complicating diagnostics. This problem has been recognized for specific genes, including many involved in human disease, and diagnostic labs must perform additional costly steps to guarantee accurate diagnosis in these cases. Here we report a new data analysis method based on the comparison of read depth between highly homologous regions to identify misalignment. Analyzing six clinically important genes—CYP21A2, GBA, HBA1/2, PMS2, and SMN1—each exhibiting misalignment issues related to homology, we show that our technique can correctly identify potential misalignment events and be used to make appropriate calls. Combined with long-range PCR and/or MLPA orthogonal testing, our clinical laboratory can improve variant calling with minimal additional cost. We propose an accurate and cost-efficient NGS testing procedure that will benefit disease diagnostics, carrier screening, and research-based population studies.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Bailey JA, Yavor AM, Massa HF et al (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017. https://doi.org/10.1101/gr.gr-1871r
Blount J, Prakash A (2018) The changing landscape of Lynch syndrome due to PMS2 mutations. Clin Genet 94:61–69. https://doi.org/10.1111/cge.13205
Butchbach MER (2016) Copy number variations in the survival motor neuron genes: implications for spinal muscular atrophy and other neurodegenerative diseases. Front Mol Biosci 3:7. https://doi.org/10.3389/fmolb.2016.00007
Campbell L, Potter A, Ignatius J et al (1997) Genomic variation and gene conversion in spinal muscular atrophy: implications for disease process and clinical phenotype. Am J Hum Genet 61:40–50. https://doi.org/10.1086/513886
Chen W, Xu Z, Sullivan A et al (2012) Junction site analysis of chimeric CYP21A1P/CYP21A2 genes in 21-hydroxylase deficiency. Clin Chem 58:421–430. https://doi.org/10.1373/clinchem.2011.174037
Choi J-H, Kim G-H, Yoo H-W (2016) Recent advances in biochemical and molecular analysis of congenital adrenal hyperplasia due to 21-hydroxylase deficiency. Ann Pediatr Endocrinol Metab 21:1–6. https://doi.org/10.6065/apem.2016.21.1.1
Concolino P, Mello E, Minucci A et al (2009) A new CYP21A1P/CYP21A2 chimeric gene identified in an Italian woman suffering from classical congenital adrenal hyperplasia form. BMC Med Genet 10:72. https://doi.org/10.1186/1471-2350-10-72
David M, Forest MG (1984) Prenatal treatment of congenital adrenal hyperplasia resulting from 21-hydroxylase deficiency. J Pediatr 105:799–803. https://doi.org/10.1016/s0022-3476(84)80310-8
Fang P, Li L, Zeng J et al (2015) Molecular characterization and copy number of SMN1, SMN2 and NAIP in Chinese patients with spinal muscular atrophy and unrelated healthy controls. BMC Musculoskelet Disord 16:11. https://doi.org/10.1186/s12891-015-0457-x
Farashi S, Harteveld CL (2018) Molecular basis of α-thalassemia. Blood Cells Mol Dis 70:43–53. https://doi.org/10.1016/j.bcmd.2017.09.004
Gan-Or Z, Liong C, Alcalay RN (2018) GBA-associated Parkinson’s disease and other synucleinopathies. Curr Neurol Neurosci Rep 18:44. https://doi.org/10.1007/s11910-018-0860-4
Goodenberger ML, Thomas BC, Riegert-Johnson D et al (2016) PMS2 monoallelic mutation carriers: the known unknown. Genet Med 18:13–19. https://doi.org/10.1038/gim.2015.27
Gould GM, Grauman PV, Theilmann MR et al (2018) Detecting clinically actionable variants in the 3’ exons of PMS2 via a reflex workflow based on equivalent hybrid capture of the gene and its pseudogene. BMC Med Genet 19:176. https://doi.org/10.1186/s12881-018-0691-9
Gupta PK, Adamtziki E, Budde U et al (2005) Gene conversions are a common cause of von Willebrand disease. Br J Haematol 130:752–758. https://doi.org/10.1111/j.1365-2141.2005.05660.x
Hasan MS, Wu X, Zhang L (2019) Uncovering missed indels by leveraging unmapped reads. Sci Rep 9:. https://doi.org/10.1038/s41598-019-47405-z
Hsieh P, Yamane K (2008) DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev 129:391–407. https://doi.org/10.1016/j.mad.2008.02.012
Innan H, Kondrashov F (2010) The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 11:97–108. https://doi.org/10.1038/nrg2689
Jia H, Guo Y, Zhao W, Wang K (2014) Long-range PCR in next-generation sequencing: comparison of six enzymes and evaluation on the MiSeq sequencer. Sci Rep 4:5737. https://doi.org/10.1038/srep05737
Keck W (1990) Rotatory evoked cortical potentials in normal subjects and patients with unilateral and bilateral vestibular loss. Eur Arch Otorhinolaryngol 247:222–225
Kohlmann W, Gruber SB (2004) [Updated 2018 Apr 12] Lynch syndrome. In: Adam MP, Ardinger HH, Pagon RA, et al. (eds). GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. https://www.ncbi.nlm.nih.gov/books/NBK1211/
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. https://doi.org/10.1186/gb-2009-10-3-r25
Lee H-H (2014) Mutational analysis of CYP21A2 gene and CYP21A1P pseudogene: long-range PCR on genomic DNA. Methods Mol Biol 1167:275–287. https://doi.org/10.1007/978-1-4939-0835-6_19
Lefebvre S, Bürglen L, Reboullet S et al (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell 80:155–165. https://doi.org/10.1016/0092-8674(95)90460-3
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. https://doi.org/10.1093/bioinformatics/btp324
Li J, Dai H, Feng Y et al (2015) A comprehensive strategy for accurate mutation detection of the highly homologous PMS2. J Mol Diagn 17:545–553. https://doi.org/10.1016/j.jmoldx.2015.04.001
Magadum S, Banerjee U, Murugan P et al (2013) Gene duplication as a major force in evolution. J Genet 92:155–161. https://doi.org/10.1007/s12041-013-0212-8
Mandelker D, Schmidt RJ, Ankala A et al (2016) Navigating highly homologous genes in a molecular diagnostic setting: a resource for clinical next-generation sequencing. Genet Med 18:1282–1289. https://doi.org/10.1038/gim.2016.58
Masurel-Paulet A, Andrieux J, Callier P et al (2010) Delineation of 15q13.3 microdeletions. Clin Genet 78:149–161. https://doi.org/10.1111/j.1399-0004.2010.01374.x
New MI, Abraham M, Yuen T, Lekarev O (2012) An update on prenatal diagnosis and treatment of congenital adrenal hyperplasia. Semin Reprod Med 30:396–399. https://doi.org/10.1055/s-0032-1324723
Nimkarn S, Gangishetti PK, Yau M, New MI (2002) [Updated 2016 Feb 4] 21-Hydroxylase-deficient congenital adrenal hyperplasia. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. https://www.ncbi.nlm.nih.gov/books/NBK1171/
Origa R, Moi P (2005) [Updated 2016 Dec 29] Alpha-Thalassemia. In: Adam MP, Ardinger HH, Pagon RA, et al. (eds) GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. https://www.ncbi.nlm.nih.gov/books/NBK1435/
Pastores GM, Hughes DA (2000) [Updated 2018 Jun 21] Gaucher Disease. In: Adam MP, Ardinger HH, Pagon RA, et al. (eds) GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. https://www.ncbi.nlm.nih.gov/books/NBK1435/
Piel FB, Weatherall DJ (2014) The α-thalassemias. N Engl J Med 371:1908–1916. https://doi.org/10.1056/NEJMra1404415
Prasun P, Hankerd M, Kristofice M et al (2014) Compound heterozygous microdeletion of chromosome 15q13.3 region in a child with hypotonia, impaired vision, and global developmental delay. Am J Med Genet A 164A:1815–1820. https://doi.org/10.1002/ajmg.a.36535
Prior TW, Finanger E (2000) [Updated 2019 Nov 14] Spinal muscular atrophy. In: Adam MP, Ardinger HH, Pagon RA et al. (eds) GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2020. https://www.ncbi.nlm.nih.gov/books/NBK1352/
Richards S, Aziz N, Bale S et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet Med 17:405–424. https://doi.org/10.1038/gim.2015.30
Smith TF, Waterman MS (1981) Identification of common molecular subsequences. J Mol Biol 147:195–197. https://doi.org/10.1016/0022-2836(81)90087-5
Thankaswamy-Kosalai S, Sen P, Nookaew I (2017) Evaluation and assessment of read-mapping by multiple next-generation sequencing aligners based on genome-wide characteristics. Genomics 109:186–191. https://doi.org/10.1016/j.ygeno.2017.03.001
Thauvin-Robinet C, Drunat S, Saugier Veber P et al (2012) Homozygous SMN1 exons 1–6 deletion: pitfalls in genetic counseling and general recommendations for spinal muscular atrophy molecular diagnosis. Am J Med Genet A 158A:1735–1741. https://doi.org/10.1002/ajmg.a.35402
Xu Z, Chen W, Merke DP, McDonnell NB (2013) Comprehensive mutation analysis of the CYP21A2 gene: an efficient multistep approach to the molecular diagnosis of congenital adrenal hyperplasia. J Mol Diagn 15:745–753. https://doi.org/10.1016/j.jmoldx.2013.06.001
Zampieri S, Cattarossi S, Bembi B, Dardis A (2017) GBA Analysis in next-generation era: pitfalls, challenges, and possible solutions. J Mol Diagn 19:733–741. https://doi.org/10.1016/j.jmoldx.2017.05.005
Zheleznyakova GY, Kiselev AV, Vakharlovsky VG et al (2011) Genetic and expression studies of SMN2 gene in Russian patients with spinal muscular atrophy type II and III. BMC Med Genet 12:96. https://doi.org/10.1186/1471-2350-12-96
Zheng GX, Lau BT, Schnall-Levin M et al (2016) Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat Biotechnol 34:303–311. https://doi.org/10.1038/nbt.3432
Acknowledgements
We would like to show our gratitude to Dr. Yiping Shen and Dr. Samuel Strom, who provided expertise and insightful comments that improved the manuscript. We thank Dr. Kyle Proffitt for editing and improving the manuscript and references. We also thank Dr. Becky Tsai for organizing the review process. This study was supported by Fulgent Genetics, Inc.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
C-YY and H-YY proposed the model and reviewed the clinical data. AZ conducted the experiments. HG provided suggestions on research goals and supervised the project.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Ethics approval
Not applicable.
Consent to participate
Whole blood or buccal cells were obtained from the subjects subsequent to informed consent.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, Cy., Yen, HY., Zhong, A.W. et al. Resolving misalignment interference for NGS-based clinical diagnostics. Hum Genet 140, 477–492 (2021). https://doi.org/10.1007/s00439-020-02216-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02216-5