Abstract
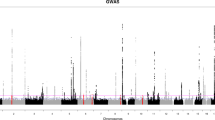
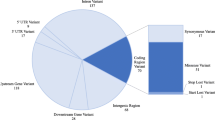
Genome-wide association studies (GWAS) have identified hundreds of primarily non-coding disease-susceptibility variants that further need functional interpretation to prioritize and discriminate the disease-relevant variants. We present a comprehensive genome-wide non-coding variant prioritization scheme followed by validation using Pyrosequencing and TaqMan assays in asthma. We implemented a composite Functional Annotation Score (cFAS) to investigate over 32,000 variants consisting of 1525 GWAS-lead asthma-susceptibility variants and their LD proxies (r2 ≥ 0.80). Functional annotation pipeline in cFAS revealed 274 variants with significant score at 1% false discovery rate. This study implicates a novel locus 4p16 (SLC26A1) with eQTL variant (rs11936407) and known loci in 17q12-21 and 5q22 which encode ORM1-like protein 3 (ORMDL3, rs406527, and rs12936231) and thymic stromal lymphopoietin (TSLP, rs3806932 and rs10073816) epithelial gene, respectively. Follow-up validation analysis through pyrosequencing of CpG sites in and nearby rs4065275 and rs11936407 showed genotype-dependent hypomethylation on asthma cases compared with healthy controls. Prioritized variants are enriched for asthma-specific histone modification associated with active chromatin (H3K4me1 and H3K27ac) in T cells, B cells, lung, and immune-related interferon gamma signaling pathways. Our findings, together with those from prior studies, suggest that SNPs can affect asthma by regulating enhancer activity, and our comprehensive bioinformatics and functional analysis could lead to biological insights into asthma pathogenesis.
Graphic abstract








Similar content being viewed by others
Change history
04 May 2020
In the original article published, the “<Emphasis Type="Italic">p</Emphasis>” value in the Fig. 5 legend is incorrectly presented as *<Emphasis Type="Italic">p </Emphasis>< 0.50. The correct <Emphasis Type="Italic">p</Emphasis> value is <Emphasis Type="Italic">*p</Emphasis> < 0.050.
References
Barnes KC (2011) Genetic studies of the etiology of asthma. Proc Am Thorac Soc 8:143–148. https://doi.org/10.1513/pats.201103-030MS
Barnes PJ, Adcock IM (1998) Transcription factors and asthma. Eur Respir J 12:221–234
Baye TM et al (2011) Differences in candidate gene association between European ancestry and African American asthmatic children. PLoS ONE 6:e16522. https://doi.org/10.1371/journal.pone.0016522
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B (Methodological) 57:298–300
Boyle AP et al (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797. https://doi.org/10.1101/gr.137323.112
Buniello A et al (2019) (2019) The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics. Nucleic Acids Res 47:D1005–D1012. https://doi.org/10.1093/nar/gky1120
Butsch Kovacic M et al (2012) The greater cincinnati pediatric clinic repository: a novel framework for childhood asthma and allergy research. Pediatr Allergy Immunol Pulmonol 25:104–113. https://doi.org/10.1089/ped.2011.0116
Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R (2010) The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet 19:111–121. https://doi.org/10.1093/hmg/ddp471
Chan IH et al (2009) Association of early growth response-1 gene polymorphisms with total IgE and atopy in asthmatic children. Pediatr Allergy Immunol 20:142–150. https://doi.org/10.1111/j.1399-3038.2008.00757.x
Chen G et al (2015) Re-annotation of presumed noncoding disease/trait-associated genetic variants by integrative analyses. Sci Rep 5:9453. https://doi.org/10.1038/srep09453
Coetzee GA, Jia L, Frenkel B, Henderson BE, Tanay A, Haiman CA, Freedman ML (2010) A systematic approach to understand the functional consequences of non-protein coding risk regions. Cell Cycle 9:256–259. https://doi.org/10.4161/cc.9.2.10419
Consortium EP (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. https://doi.org/10.1038/nature11247
Cotsapas C et al (2011) Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 7:e1002254. https://doi.org/10.1371/journal.pgen.1002254
Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S (2010) Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6:e1001025. https://doi.org/10.1371/journal.pcbi.1001025
Demenais F et al (2018) Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet 50:42–53. https://doi.org/10.1038/s41588-017-0014-7
Dimas AS et al (2009) Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325:1246–1250. https://doi.org/10.1126/science.1174148
Fahy JV (2009) Eosinophilic and neutrophilic inflammation in asthma: insights from clinical studies. Proc Am Thorac Soc 6:256–259. https://doi.org/10.1513/pats.200808-087RM
Freedman ML et al (2011) Principles for the post-GWAS functional characterization of cancer risk loci. Nat Genet 43:513–518. https://doi.org/10.1038/ng.840
Garber M, Guttman M, Clamp M, Zody MC, Friedman N, Xie X (2009) Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25:i54–i62. https://doi.org/10.1093/bioinformatics/btp190
Garrett-Sinha LA (2013) Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci 70:3375–3390. https://doi.org/10.1007/s00018-012-1243-7
Gashler A, Sukhatme VP (1995) Early growth response protein 1 (Egr-1): prototype of a zinc-finger family of transcription factors. Prog Nucleic Acid Res Mol Biol 50:191–224
Gauvreau GM et al (2014) Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med 370:2102–2110. https://doi.org/10.1056/NEJMoa1402895
Genomes Project C et al (2015) A global reference for human genetic variation. Nature 526:68–74. https://doi.org/10.1038/nature15393
Gerasimova A et al (2013) Predicting cell types and genetic variations contributing to disease by combining GWAS and epigenetic data. PLoS ONE 8:e54359. https://doi.org/10.1371/journal.pone.0054359
Griffin MJ, Zhou Y, Kang S, Zhang X, Mikkelsen TS, Rosen ED (2013) Early B-cell factor-1 (EBF1) is a key regulator of metabolic and inflammatory signaling pathways in mature adipocytes. J Biol Chem 288:35925–35939. https://doi.org/10.1074/jbc.M113.491936
Harada M et al (2009) Functional analysis of the thymic stromal lymphopoietin variants in human bronchial epithelial cells. Am J Respir Cell Mol Biol 40:368–374. https://doi.org/10.1165/rcmb.2008-0041OC
Heintzman ND et al (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112. https://doi.org/10.1038/nature07829
Herwig R, Hardt C, Lienhard M, Kamburov A (2016) Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat Protoc 11:1889–1907. https://doi.org/10.1038/nprot.2016.117
Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 106:9362–9367. https://doi.org/10.1073/pnas.0903103106
Hnisz D et al (2013) Super-enhancers in the control of cell identity and disease. Cell 155:934–947. https://doi.org/10.1016/j.cell.2013.09.053
Jiang K, Zhu L, Buck MJ, Chen Y, Carrier B, Liu T, Jarvis JN (2015) Disease-associated single-nucleotide polymorphisms from noncoding regions in juvenile idiopathic arthritis are located within or adjacent to functional genomic elements of human neutrophils and CD4+ T cells. Arthritis Rheumatol 67:1966–1977. https://doi.org/10.1002/art.39135
Karwot R et al (2008) Protective role of nuclear factor of activated T cells 2 in CD8+ long-lived memory T cells in an allergy model. J Allergy Clin Immunol 121:992–999e996. https://doi.org/10.1016/j.jaci.2007.12.1172
Lasky-Su J et al (2012) HLA-DQ strikes again: genome-wide association study further confirms HLA-DQ in the diagnosis of asthma among adults. Clin Exp Allergy 42:1724–1733. https://doi.org/10.1111/cea.12000
Leslie R, O'Donnell CJ, Johnson AD (2014) GRASP: analysis of genotype-phenotype results from 1390 genome-wide association studies and corresponding open access database. Bioinformatics 30:i185–i194. https://doi.org/10.1093/bioinformatics/btu273
Li MJ et al (2016) Predicting regulatory variants with composite statistic. Bioinformatics 32:2729–2736. https://doi.org/10.1093/bioinformatics/btw288
Li B, Lu Q, Zhao H (2017) An evaluation of noncoding genome annotation tools through enrichment analysis of 15 genome-wide association studies. Brief Bioinform. https://doi.org/10.1093/bib/bbx131
Liu L, Jin G, Zhou X (2015) Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res 43:3873–3885. https://doi.org/10.1093/nar/gkv255
Lonsdale J et al (2013) The genotype-tissue expression (GTEx) project. Nat Genet 45:580–585. https://doi.org/10.1038/ng.2653
Lu Q et al (2017) Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer's disease. PLoS Genet 13:e1006933. https://doi.org/10.1371/journal.pgen.1006933
MacArthur J et al (2017) The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45:D896–D901. https://doi.org/10.1093/nar/gkw1133
Machiela MJ, Chanock SJ (2015) LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 31:3555–3557. https://doi.org/10.1093/bioinformatics/btv402
Mailman MD et al (2007) The NCBI dbGaP database of genotypes and phenotypes. Nat Genet 39:1181–1186. https://doi.org/10.1038/ng1007-1181
Maurano MT et al (2012) Systematic localization of common disease-associated variation in regulatory DNA. Science 337:1190–1195. https://doi.org/10.1126/science.1222794
Moffatt MF et al (2010) A large-scale, consortium-based genomewide association study of asthma. N Engl J Med 363:1211–1221. https://doi.org/10.1056/NEJMoa0906312
Movahedi M et al (2008) Association of HLA class II alleles with childhood asthma and total IgE levels. Iran J Allergy Asthma Immunol 7:215–220 (doi:07.04/ijaai.215220)
Pickrell JK, Berisa T, Liu JZ, Segurel L, Tung JY, Hinds DA (2016) Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 48:709–717. https://doi.org/10.1038/ng.3570
Prasad MA et al (2015) Ebf1 heterozygosity results in increased DNA damage in pro-B cells and their synergistic transformation by Pax5 haploinsufficiency. Blood 125:4052–4059. https://doi.org/10.1182/blood-2014-12-617282
Purcell S et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Ramos EM et al (2014) Phenotype-genotype integrator (PheGenI): synthesizing genome-wide association study (GWAS) data with existing genomic resources. Eur J Hum Genet 22:144–147. https://doi.org/10.1038/ejhg.2013.96
Ritchie GR, Dunham I, Zeggini E, Flicek P (2014) Functional annotation of noncoding sequence variants. Nat Methods 11:294–296. https://doi.org/10.1038/nmeth.2832
Roadmap Epigenomics C et al (2015) Integrative analysis of 111 reference human epigenomes. Nature 518:317–330. https://doi.org/10.1038/nature14248
Russell L, Garrett-Sinha LA (2010) Transcription factor Ets-1 in cytokine and chemokine gene regulation. Cytokine 51:217–226. https://doi.org/10.1016/j.cyto.2010.03.006
Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M (2012) Linking disease associations with regulatory information in the human genome. Genome Res 22:1748–1759. https://doi.org/10.1101/gr.136127.111
Schmiedel BJ et al (2016) 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun 7:13426. https://doi.org/10.1038/ncomms13426
Shi H, Kichaev G, Pasaniuc B (2016) Contrasting the genetic architecture of 30 complex traits from summary association data. Am J Hum Genet 99:139–153. https://doi.org/10.1016/j.ajhg.2016.05.013
Silverman ES et al (2001) The transcription factor early growth-response factor 1 modulates tumor necrosis factor-alpha, immunoglobulin E, and airway responsiveness in mice. Am J Respir Crit Care Med 163:778–785. https://doi.org/10.1164/ajrccm.163.3.2003123
Stein MM et al (2018) A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle. J Allergy Clin Immunol. https://doi.org/10.1016/j.jaci.2017.12.974
Torgerson DG et al (2011) Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet 43:887–892. https://doi.org/10.1038/ng.888
Trynka G, Sandor C, Han B, Xu H, Stranger BE, Liu XS, Raychaudhuri S (2013) Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat Genet 45:124–130. https://doi.org/10.1038/ng.2504
Vicente CT, Revez JA, Ferreira MAR (2017) Lessons from ten years of genome-wide association studies of asthma. Clin Transl Immunol 6:e165. https://doi.org/10.1038/cti.2017.54
Wang K, Li M, Hakonarson H (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. https://doi.org/10.1093/nar/gkq603
Wang L, Oehlers SH, Espenschied ST, Rawls JF, Tobin DM, Ko DC (2015) CPAG: software for leveraging pleiotropy in GWAS to reveal similarity between human traits links plasma fatty acids and intestinal inflammation. Genome Biol 16:190. https://doi.org/10.1186/s13059-015-0722-1
Ward LD, Kellis M (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 44:D877–D881. https://doi.org/10.1093/nar/gkv1340
Zahran HS, Bailey C, Garbe P (2011) Vital signs: asthma prevalence, disease characteristics, and self-management education—United States, 2001–2009, vol 60
Acknowledgements
This work was supported by the National Institutes of Health (NIH) Grant R01HL132344. The CAMP/CARE and STAMPEED datasets were obtained from dbGap through accession number phs000166.v2.p1 and phs000355.v1.p1, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gautam, Y., Afanador, Y., Ghandikota, S. et al. Comprehensive functional annotation of susceptibility variants associated with asthma. Hum Genet 139, 1037–1053 (2020). https://doi.org/10.1007/s00439-020-02151-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-020-02151-5