Abstract
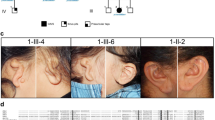
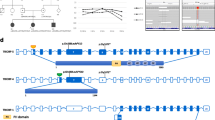
Congenital inner ear malformations affecting both the osseous and membranous labyrinth can have a devastating impact on hearing and language development. With the exception of an enlarged vestibular aqueduct, non-syndromic inner ear malformations are rare, and their underlying molecular biology has thus far remained understudied. To identify molecular factors that might be important in the developing inner ear, we adopted a family-based trio exome sequencing approach in young unrelated subjects with severe inner ear malformations. We identified two previously unreported de novo loss-of-function variants in GREB1L [c.4368G>T;p.(Glu1410fs) and c.982C>T;p.(Arg328*)] in two affected subjects with absent cochleae and eighth cranial nerve malformations. The cochlear aplasia in these affected subjects suggests that a developmental arrest or problem at a very early stage of inner ear development exists, e.g., during the otic pit formation. Craniofacial Greb1l RNA expression peaks in mice during this time frame (E8.5). It also peaks in the developing inner ear during E13–E16, after which it decreases in adulthood. The crucial function of Greb1l in craniofacial development is also evidenced in knockout mice, which develop severe craniofacial abnormalities. In addition, we show that Greb1l−/− zebrafish exhibit a loss of abnormal sensory epithelia innervation. An important role for Greb1l in sensory epithelia innervation development is supported by the eighth cranial nerve deficiencies seen in both affected subjects. In conclusion, we demonstrate that GREB1L is a key player in early inner ear and eighth cranial nerve development. Abnormalities in cochleovestibular anatomy can provide challenges for cochlear implantation. Combining a molecular diagnosis with imaging techniques might aid the development of individually tailored therapeutic interventions in the future.



Similar content being viewed by others
References
Aldrich J, Keats JJ, Liang WS et al (2016) Abstract 45: detection of focal somatic copy number variants in whole genome, whole exome, and targeted next-generation sequencing data of tumor/normal pairs. Clin Cancer Res 22:45–45. https://doi.org/10.1158/1557-3265.PMSCLINGEN15-45
Birman CS, Powell HRF, Gibson WPR, Elliott EJ (2016) Cochlear implant outcomes in cochlea nerve aplasia and hypoplasia. Otol Neurotol 37:438–445. https://doi.org/10.1097/MAO.0000000000000997
Boissel S, Fallet-Bianco C, Chitayat D et al (2017) Genomic study of severe fetal anomalies and discovery of GREB1L mutations in renal agenesis. Genet Med. https://doi.org/10.1038/gim.2017.173
Brophy PD, Rasmussen M, Parida M et al (2017) A gene implicated in activation of retinoic acid receptor targets is a novel renal agenesis gene in humans. Genetics 207:215–228. https://doi.org/10.1534/genetics.117.1125
Brunskill EW, Potter AS, Distasio A et al (2014) A gene expression atlas of early craniofacial development. Dev Biol 391:133–146. https://doi.org/10.1016/j.ydbio.2014.04.016
Chadha NK, James AL, Gordon KA et al (2009) Bilateral cochlear implantation in children with anomalous cochleovestibular anatomy. Arch Otolaryngol Neck Surg 135:903. https://doi.org/10.1001/archoto.2009.120
Chi F-L, Han Z, Dai P-D et al (2009) Three-dimensional reconstruction of C57BL/6 mouse inner ear during development. ORL J Otorhinolaryngol Relat Spec 71:334–341. https://doi.org/10.1159/000272029
Cingolani P, Platts A, Wang LL et al (2012) A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. https://doi.org/10.4161/fly.19695
Colletti L, Wilkinson EP, Colletti V (2013) Auditory brainstem implantation after unsuccessful cochlear implantation of children with clinical diagnosis of cochlear nerve deficiency. Ann Otol Rhinol Laryngol 122:605–612
Cryns K, Thys S, Van Laer L et al (2003) The WFS1 gene, responsible for low frequency sensorineural hearing loss and Wolfram syndrome, is expressed in a variety of inner ear cells. Histochem Cell Biol 119:247–256. https://doi.org/10.1007/s00418-003-0495-6
De Tomasi L, David P, Humbert C et al (2017) Mutations in GREB1L cause bilateral kidney agenesis in humans and mice. Am J Hum Genet 101:803–814. https://doi.org/10.1016/j.ajhg.2017.09.026
Firth HV, Richards SM, Bevan AP et al (2009) DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am J Hum Genet 84:524–533. https://doi.org/10.1016/j.ajhg.2009.03.010
Halperin RF, Carpten JD, Manojlovic Z et al (2017) A method to reduce ancestry related germline false positives in tumor only somatic variant calling. BMC Med Genom 10:61. https://doi.org/10.1186/s12920-017-0296-8
Huang N, Lee I, Marcotte EM, Hurles ME (2010) Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 6:e1001154. https://doi.org/10.1371/journal.pgen.1001154
Jackler RK, Luxford WM, House WF (1987) Congenital malformations of the inner ear: a classification based on embryogenesis. Laryngoscope 97:2–14
Jian X, Boerwinkle E, Liu X (2014) In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res 42:13534–13544. https://doi.org/10.1093/nar/gku1206
Kaga K (2017) Overview. In: Cochlear implantation in children with inner ear malformation and cochlear nerve deficiency. Springer Singapore, Singapore, pp 1–9
Kaplan AB, Kozin ED, Puram SV et al (2015) Auditory brainstem implant candidacy in the United States in children 0–17 years old. Int J Pediatr Otorhinolaryngol 79:310–315. https://doi.org/10.1016/j.ijporl.2014.11.023
Kari E, Schrauwen I, Llaci L et al (2017) Compound heterozygous mutations in MASP1 in a deaf child with absent cochlear nerves. Neurol Genet 3:e153. https://doi.org/10.1212/NXG.0000000000000153
Kari E, Go JL, Loggins J et al (2018) Abnormal cochleovestibular nerves and pediatric hearing outcomes: patients with “absent cochlear nerves” can derive benefit from cochlear implantation. Otol Neurotol (in press)
Kircher M, Witten DM, Jain P et al (2014) A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 46:310–315. https://doi.org/10.1038/ng.2892
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. https://doi.org/10.1186/gb-2009-10-3-r25
Lek M, Karczewski KJ, Minikel EV et al (2016) Analysis of protein-coding genetic variation in 60,706 humans. Nature 536:285–291. https://doi.org/10.1038/nature19057
Liu H, Pecka JL, Zhang Q et al (2014) Characterization of transcriptomes of cochlear inner and outer hair cells. J Neurosci 34:11085–11095. https://doi.org/10.1523/JNEUROSCI.1690-14.2014
Liu X, Wu C, Li C, Boerwinkle E (2016) dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat 37:235–241. https://doi.org/10.1002/humu.22932
Lu CC, Appler JM, Houseman EA, Goodrich LV (2011) Developmental profiling of spiral ganglion neurons reveals insights into auditory circuit assembly. J Neurosci 31:10903–10918. https://doi.org/10.1523/JNEUROSCI.2358-11.2011
MacDonald JR, Ziman R, Yuen RKC et al (2014) The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res 42:D986–D992. https://doi.org/10.1093/nar/gkt958
Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5:89–99. https://doi.org/10.1038/nrm1310
Marques AH, O’Connor TG, Roth C et al (2013) The influence of maternal prenatal and early childhood nutrition and maternal prenatal stress on offspring immune system development and neurodevelopmental disorders. Front Neurosci 7:120. https://doi.org/10.3389/fnins.2013.00120
Masuda S, Usui S, Matsunaga T (2013) High prevalence of inner-ear and/or internal auditory canal malformations in children with unilateral sensorineural hearing loss. Int J Pediatr Otorhinolaryngol 77:228–232. https://doi.org/10.1016/j.ijporl.2012.11.001
McKenna A, Hanna M, Banks E et al (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. https://doi.org/10.1101/gr.107524.110
Orten DJ, Fischer SM, Sorensen JL et al (2008) Branchio-oto-renal syndrome (BOR): novel mutations in the EYA1 gene, and a review of the mutational genetics of BOR. Hum Mutat 29:537–544. https://doi.org/10.1002/humu.20691
Pai I (2017) Embryology of cochlear nerve and its deficiency. In: Kaga K (ed) Cochlear implantation in children with inner ear malformation and cochlear nerve deficiency. Springer Singapore, Singapore, pp 19–27
Plouhinec J-L, Roche DD, Pegoraro C et al (2014) Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev Biol 386:461–472. https://doi.org/10.1016/j.ydbio.2013.12.010
Pryor SP (2005) SLC26A4/PDS genotype–phenotype correlation in hearing loss with enlargement of the vestibular aqueduct (EVA): evidence that Pendred syndrome and non-syndromic EVA are distinct clinical and genetic entities. J Med Genet 42:159–165. https://doi.org/10.1136/jmg.2004.024208
Rae JM, Johnson MD, Scheys JO et al (2005) GREB1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat 92:141–149. https://doi.org/10.1007/s10549-005-1483-4
Raible DW, Kruse GJ (2000) Organization of the lateral line system in embryonic zebrafish. J Comp Neurol 421:189–198
Rooryck C, Diaz-Font A, Osborn DPS et al (2011) Mutations in lectin complement pathway genes COLEC11 and MASP1 cause 3MC syndrome. Nat Genet 43:197–203. https://doi.org/10.1038/ng.757
Sandell LL, Butler Tjaden NE, Barlow AJ, Trainor PA (2014) Cochleovestibular nerve development is integrated with migratory neural crest cells. Dev Biol 385:200–210. https://doi.org/10.1016/j.ydbio.2013.11.009
Sanna-Cherchi S, Khan K, Westland R et al (2017) Exome-wide association study identifies GREB1L mutations in congenital kidney malformations. Am J Hum Genet 101:789–802. https://doi.org/10.1016/j.ajhg.2017.09.018
Scheffer DI, Shen J, Corey DP, Chen Z-Y (2015) Gene Expression by mouse inner ear hair cells during development. J Neurosci 35:6366–6380. https://doi.org/10.1523/JNEUROSCI.5126-14.2015
Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ et al (2016) A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res. https://doi.org/10.1016/j.heares.2015.08.013
Sennaroglu L, Bajin MD (2017) Classification of inner ear malformations. In: Kaga K (ed) Cochlear implantation in children with inner ear malformation and cochlear nerve deficiency. Springer Singapore, Singapore, pp 61–85
Sennaroglu L, Saatci I (2002) A new classification for cochleovestibular malformations. Laryngoscope 112:2230–2241. https://doi.org/10.1097/00005537-200212000-00019
Shen J, Scheffer DI, Kwan KY, Corey DP (2015) SHIELD: an integrative gene expression database for inner ear research. Database (Oxford) 2015:bav071. https://doi.org/10.1093/database/bav071
Torres M, Giráldez F (1998) The development of the vertebrate inner ear. Mech Dev 71:5–21. https://doi.org/10.1016/S0925-4773(97)00155-X
Westerfield M (1993) The zebrafish book: a guide for the laboratory use of zebrafish, 2nd edn. University of Oregon Press, Eugene
Xu P-X, Zheng W, Laclef C et al (2002) Eya1 is required for the morphogenesis of mammalian thymus, parathyroid and thyroid. Development 129:3033–3044
Yamamoto N, Kanno A, Matsunaga T (2017) Genetics of inner ear malformation and cochlear nerve deficiency. In: Kaga K (ed) Cochlear implantation in children with inner ear malformation and cochlear nerve deficiency. Springer Singapore, Singapore, pp 47–59
Yang Q, Sun P, Chen S et al (2017) Behavioral methods for the functional assessment of hair cells in zebrafish. Front Med 11:178–190. https://doi.org/10.1007/s11684-017-0507-x
Yates JA, Patel PC, Millman B, Gibson WS (1997) Isolated congenital internal auditory canal atresia with normal facial nerve function. Int J Pediatr Otorhinolaryngol 41:1–8
Ye S, Dhillon S, Ke X et al (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29:E88–E88
Young NM, Kim FM, Ryan ME et al (2012) Pediatric cochlear implantation of children with eighth nerve deficiency. Int J Pediatr Otorhinolaryngol 76:1442–1448. https://doi.org/10.1016/j.ijporl.2012.06.019
Acknowledgements
The authors thank the families for participating in this study. This study was supported by private donations to TGen’s Center for Rare Childhood Disorders (https://www.tgen.org/giving/tgen-foundation/), the American Hearing Research Foundation to I.S. (http://american-hearing.org/), National Institutes of Health R01 010856 (https://www.nih.gov/) and the Mills Auditory Foundation (http://millsauditoryfoundation.org/) to R.A.F. We would like to acknowledge Brunskill et al., Lu et al., Liu et al., and Scheffer et al. for the creation and public deposition of their RNA expression data that was used in this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
Institutional review board (IRB) approval for human research was obtained, and the principles outlined in the Declaration of Helsinki were followed.
Informed consent
Informed consent was obtained from the participants involved (University of Southern California (USC) IRB #HS-14-00513-CR002 and Western (IRB) #20120512). The Institutional Animal Care and Use Committee of the USC approved the animal experiments performed in this study (no. 10885).
Conflict of interest
The authors declare that they have no conflict of interest.
Web resources
Bravo TOPMed variant browser, https://bravo.sph.umich.edu/. Burrows-Wheeler Aligner, http://bio-bwa.sourceforge.net/. CDC, hearing loss in children, http://cdc.gov/ncbddd/hearingloss/data.html/. Clinvar, https://www.ncbi.nlm.nih.gov/clinvar/. Combined Annotation Dependent Depletion (CADD), http://cadd.gs.washington.edu/. dbNSFP, https://sites.google.com/site/jpopgen/dbNSFP/. dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/. Database of Genomic Variants (DGV), http://dgv.tcag.ca/dgv/app/home/. DatabasE of genomiC varIation and Phenotype in Humans using Ensembl Resources (DECIPHER), https://decipher.sanger.ac.uk/. Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org/. Genome Aggregation Database (gnomAD), http://gnomad.broadinstitute.org/. Genome Analysis Toolkit (GATK), https://software.broadinstitute.org/gatk/. Genome Browser, https://genome.ucsc.edu/. Online Mendelian Inheritance of Man (OMIM), https://www.omim.org/. Picard, http://broadinstitute.github.io/picard/.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Schrauwen, I., Kari, E., Mattox, J. et al. De novo variants in GREB1L are associated with non-syndromic inner ear malformations and deafness. Hum Genet 137, 459–470 (2018). https://doi.org/10.1007/s00439-018-1898-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-018-1898-8