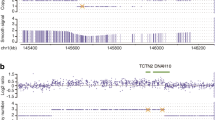
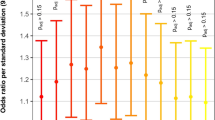
Abstract
Classic heterotaxy consists of congenital heart defects with abnormally positioned thoracic and abdominal organs. We aimed to uncover novel, genomic copy-number variants (CNVs) in classic heterotaxy cases. A microarray containing 2.5 million single-nucleotide polymorphisms (SNPs) was used to genotype 69 infants (cases) with classic heterotaxy identified from California live births from 1998 to 2009. CNVs were identified using the PennCNV software. We identified 56 rare CNVs encompassing genes in the NODAL (NIPBL, TBX6), BMP (PPP4C), and WNT (FZD3) signaling pathways, not previously linked to classic heterotaxy. We also identified a CNV involving FGF12, a gene previously noted in a classic heterotaxy case. CNVs involving RBFOX1 and near MIR302F were detected in multiple cases. Our findings illustrate the importance of body patterning pathways for cardiac development and left/right axes determination. FGF12, RBFOX1, and MIR302F could be important in human heterotaxy, because they were noted in multiple cases. Further investigation into genes involved in the NODAL, BMP, and WNT body patterning pathways and into the dosage effects of FGF12, RBFOX1, and MIR302F is warranted.
Similar content being viewed by others
References
Campos CM, Zanardo EA, Dutra RL, Kulikowski LD, Kim CA (2015) Investigation of copy number variation in children with conotruncal heart defects. Arq Bras Cardiol 104:24–31. doi:10.5935/abc.20140169
Cohen PT, Philp A, Vazquez-Martin C (2005) Protein phosphatase 4–from obscurity to vital functions. FEBS Lett 579:3278–3286. doi:10.1016/j.febslet.2005.04.070
Croen LA, Shaw GM, Jensvold NG, Harris JA (1991) Birth defects monitoring in California: a resource for epidemiological research. Paediatr Perinat Epidemiol 5:423–427. doi:10.1111/j.1365-3016.1991.tb00728.x
Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M (2011) Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc Natl Acad Sci 108:2915–2920. doi:10.1073/pnas.1019645108
Feng Y, Wu H, Xu Y, Zhang Z, Liu T, Lin X, Feng XH (2014) Zinc finger protein 451 is a novel Smad corepressor in transforming growth factor-beta signaling. J Biol Chem 289:2072–2083. doi:10.1074/jbc.M113.526905
Ferencz C, Loffredo CA, Correa-Villaseñor A, Wilson PD (1997) Genetic and environmental risk factors of major cardiovascular malformations. In: The Baltimore-Washington infant study 1981–1989, perspectives in pediatric cardiology, vol V. Futura Publishing, Armonk
Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG (2011) Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol 359:251–261. doi:10.1016/j.ydbio.2011.08.025
Geng J, Picker J, Zheng Z, Zhang X, Wang J, Hisama F, Brown DW, Mullen MP, Harris D, Stoler J, Seman A, Miller DT, Fu Q, Roberts AE, Shen Y (2014) Chromosome microarray testing for patients with congenital heart defects reveals novel disease causing loci and high diagnostic yield. BMC Genom 15:1127. doi:10.1186/1471-2164-15-1127
Ghebranious N, Giampietro PF, Wesbrook FP, Rezkalla SH (2007) A novel microdeletion at 16p11.2 harbors candidate genes for aortic valve development, seizure disorder, and mild mental retardation. Am J Med Genet A 143A:1462–1471. doi:10.1002/ajmg.a.31837
Hadjantonakis AK, Pisano E, Papaioannou VE (2008) Tbx6 regulates left/right patterning in mouse embryos through effects on nodal cilia and perinodal signaling. PLoS One 3:e2511. doi:10.1371/journal.pone.0002511
Hartung H, Feldman B, Lovec H, Coulier F, Birnbaum D, Goldfarb M (1997) Murine FGF-12 and FGF-13: expression in embryonic nervous system, connective tissue and heart. Mech Dev 64:31–39. doi:10.1016/S0925-4773(97)00042-7
Jia S, Dai F, Wu D, Lin X, Xing C, Xue Y, Wang Y, Xiao M, Wu W, Feng XH, Meng A (2012) Protein phosphatase 4 cooperates with Smads to promote BMP signaling in dorsoventral patterning of zebrafish embryos. Dev Cell 22:1065–1078. doi:10.1016/j.devcel.2012.03.001
Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, Ahrens P, Lange L, Morillas HN, Noone PG, Zariwala MA, Knowles MR (2007) Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation 115:2814–2821. doi:10.1161/circulationaha.106.649038
Komatsu Y, Mishina Y (2013) Establishment of left-right asymmetry in vertebrate development: the node in mouse embryos. Cell Mol Life Sci 70:4659–4666. doi:10.1007/s00018-013-1399-9
Kurkowiak M, Ziętkiewicz E, Witt M (2015) Recent advances in primary ciliary dyskinesia genetics. J Med Genet 52(1):1–9. doi:10.1136/jmedgenet-2014-102755
Kuroyanagi H (2009) Fox-1 family of RNA-binding proteins. Cell Mol Life Sci 66:3895–3907. doi:10.1007/s00018-009-0120-5
Lale S, Yu S, Ahmed A (2011) Complex congenital heart defects in association with maternal diabetes and partial deletion of the A2BP1 gene. Fetal Pediatr Pathol 30:161–166. doi:10.3109/15513815.2010.547555
Li D, Tekin M, Buch M, Fan YS (2012) Co-existence of other copy number variations with 22q11.2 deletion or duplication: a modifier for variable phenotypes of the syndrome? Mol Cytogenet 5:18. doi:10.1186/1755-8166-5-18
Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, Tariq M, King SM, Hendricks G, Cui C, Saydmohammed M, Lee DM, Zahid M, Sami I, Leatherbury L, Pazour GJ, Ware SM, Nakanishi T, Goldmuntz E, Tsang M, Lo CW (2016) DNAH6 and its interactions with PCD genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet 12(2). doi:10.1371/journal.pgen.1005821
Lin AE, Krikov S, Riehle-Colarusso T, Frias JL, Belmont J, Anderka M, Geva T, Getz KD, Botto LD (2014) Laterality defects in the national birth defects prevention study (1998–2007): birth prevalence and descriptive epidemiology. Am J Med Genet A 164A:2581–2591. doi:10.1002/ajmg.a.36695
Lobo J, Zariwala MA, Noone PG (2015) Primary ciliary dyskinesia. Semin Respir Crit Care Med 36(2):169–179. doi:10.1055/s-0035-1546748
Mizoguchi T, Izawa T, Kuroiwa A, Kikuchi Y (2006) Fgf signaling negatively regulates Nodal-dependent endoderm induction in zebrafish. Dev Biol 300:612–622. doi:10.1016/j.ydbio.2006.08.073
Mohapatra B, Casey B, Li H, Ho-Dawson T, Smith L, Fernbach SD, Molinari L, Niesh SR, Jefferies JL, Craigen WJ, Towbin JA, Belmont JW, Ware SM (2009) Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet 18:861–871. doi:10.1093/hmg/ddn411
Muto A, Calof AL, Lander AD, Schilling TF (2011) Multifactorial origins of heart and gut defects in nipbl-deficient zebrafish, a model of Cornelia de Lange Syndrome. PLoS Biol 9:e1001181. doi:10.1371/journal.pbio.1001181
Nakano N, Maeyama K, Sakata N, Itoh F, Akatsu R, Nakata M, Katsu Y, Ikeno S, Togawa Y, Vo Nguyen TT, Watanabe Y, Kato M, Itoh S (2014) C18 ORF1, a novel negative regulator of transforming growth factor-beta signaling. J Biol Chem 289:12680–12692. doi:10.1074/jbc.M114.558981
Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, Sabol SL, Swisher M, Connelly PS, Daniels MP, Srinivasan A, Kuehl K, Kravitz N, Burns K, Sami I, Omran H, Barmada M, Olivier K, Chawla KK, Leigh M, Jonas R, Knowles M, Leatherbury L, Lo CW (2012) High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation 125(18):2232–2242. doi:10.1161/CIRCULATIONAHA.111.079780
Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ (2009) FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 458:651–654. doi:10.1038/nature07753
Rigler SL, Kay DM, Sicko RJ, Fan R, Liu A, Caggana M, Browne ML, Druschel CM, Romitti PA, Brody LC, Mills JL (2015) Novel copy-number variants in a population-based investigation of classic heterotaxy. Genet Med 17:348–357. doi:10.1038/gim.2014.112
Rochais F, Mesbah K, Kelly RG (2009) Signaling pathways controlling second heart field development. Circ Res 104:933–942. doi:10.1161/circresaha.109.194464
Rosa A, Spagnoli FM, Brivanlou AH (2009) The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16:517–527. doi:10.1016/j.devcel.2009.02.007
Schulman J, Hahn JA (1993) Quality control of birth defect registry data: a case study. Public Health Rep 108:91–98
Shen MM (2007) Nodal signaling: developmental roles and regulation. Development 134:1023–1034. doi:10.1242/dev.000166
Shiraishi I, Ichikawa H (2012) Human heterotaxy syndrome—from molecular genetics to clinical features, management, and prognosis. Circ J 76:2066–2075. doi:10.1253/circj.CJ-12-0957
Smith KA, Noel E, Thurlings I, Rehmann H, Chocron S, Bakkers J (2011) Bmp and nodal independently regulate lefty1 expression to maintain unilateral nodal activity during left-right axis specification in zebrafish. PLoS Genet 7:e1002289. doi:10.1371/journal.pgen.1002289
Srivastava D, Olson EN (2000) A genetic blueprint for cardiac development. Nature 407:221–226. doi:10.1038/35025190
Sutherland MJ, Ware SM (2009) Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet 151C:307–317. doi:10.1002/ajmg.c.30228
Vladar EK, Antic D, Axelrod JD (2009) Planar cell polarity signaling: the developing cell’s compass. Cold Spring Harb Perspect Biol 1:a002964. doi:10.1101/cshperspect.a002964
Yagi H, Furutani Y, Hamada H, Sasaki T, Asakawa S, Minoshima S, Ichida F, Joo K, Kimura M, Imamura S, Kamatani N, Momma K, Takao A, Nakazawa M, Shimizu N, Matsuoka R (2003) Role of TBX1 in human del22q11.2 syndrome. Lancet 362:1366–1373. doi:10.1016/S0140-6736(03)14632-6
Zhu L, Belmont JW, Ware SM (2006) Genetics of human heterotaxias. Eur J Hum Genet 14:17–25. doi:10.1038/sj.ejhg.5201506
Acknowledgments
We thank the CBDMP and the California Department of Public Health for case identification; Michael Tsai and Natalie Weir at the Minnesota Core Laboratories and the staff at the Biomedical Genomics Center Facility at the University of Minnesota for microarray genotyping; Matthew Shudt and Zhen Zhang at the Wadsworth Center Applied Genomics Technologies Core, New York State Department of Health, for next-generation sequencing; Zoe Edmunds and Katherine Keever at the Wadsworth Center, New York State Department of Health, for technical assistance; and Nathan Pankratz, University of Minnesota, and Karl G. Hill, Social Development Research Group, University of Washington, for generously sharing population B-allele frequency and GC content files for PennCNV software. The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was funded by the Intramural Research Program of the National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (Contracts HHSN275201100001I, HHSN27500005, and N01-DK-73431). Dr. Shaw was partially supported for this work by funds from CDC (5U01DD001033) and NIH (R01HL092330).
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hagen, E.M., Sicko, R.J., Kay, D.M. et al. Copy-number variant analysis of classic heterotaxy highlights the importance of body patterning pathways. Hum Genet 135, 1355–1364 (2016). https://doi.org/10.1007/s00439-016-1727-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1727-x