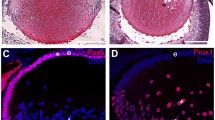
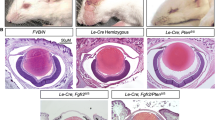
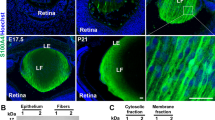
Abstract
Although majority of the genes linked to early-onset cataract exhibit lens fiber cell-enriched expression, our understanding of gene regulation in these cells is limited to function of just eight transcription factors and largely in the context of crystallins. We report on small Maf transcription factors Mafg and Mafk as regulators of several non-crystallin human cataract-associated genes in fiber cells and establish their significance to this disease. We applied a bioinformatics tool for cataract gene discovery iSyTE to identify Mafg and its co-regulators in the lens, and generated various null-allelic combinations of Mafg:Mafk mouse mutants for phenotypic and molecular analysis. By age 4 months, Mafg−/−:Mafk+/− mutants exhibit lens defects that progressively develop into cataract. High-resolution phenotypic characterization of Mafg−/−:Mafk+/− mouse lens reveals severely disorganized fiber cells, while microarray-based expression profiling identifies 97 differentially regulated genes (DRGs). Integrative analysis of Mafg−/−:Mafk+/− lens-DRGs with (1) binding motifs and genomic targets of small Mafs and their regulatory partners, (2) iSyTE lens expression data, and (3) interactions between DRGs in the String database, unravel a detailed small Maf regulatory network in the lens, several nodes of which are linked to cataract. This approach identifies 36 high-priority candidates from the original 97 DRGs. Significantly, 8/36 (22 %) DRGs are associated with cataracts in human (GSTO1, MGST1, SC4MOL, UCHL1) or mouse (Aldh3a1, Crygf, Hspb1, Pcbd1), suggesting a multifactorial etiology that includes oxidative stress and misregulation of sterol synthesis. These data identify Mafg and Mafk as new cataract-associated candidates and define their function in regulating largely non-crystallin genes linked to human cataract.







Similar content being viewed by others
References
Alizadeh A, Clark J, Seeberger T et al (2004) Characterization of a mutation in the lens-specific CP49 in the 129 strain of mouse. Invest Ophthalmol Vis Sci 45:884–891
Bassnett S, Shi Y, Vrensen GFJM (2011) Biological glass: structural determinants of eye lens transparency. Philos Trans R Soc Lond B Biol Sci 366:1250–1264. doi:10.1098/rstb.2010.0302
Bayle JH, Randazzo F, Johnen G et al (2002) Hyperphenylalaninemia and impaired glucose tolerance in mice lacking the bifunctional DCoH gene. J Biol Chem 277:28884–28891. doi:10.1074/jbc.M201983200
Berry V, Francis P, Reddy MA et al (2001) Alpha-B crystallin gene (CRYAB) mutation causes dominant congenital posterior polar cataract in humans. Am J Hum Genet 69:1141–1145. doi:10.1086/324158
Berthoud VM, Beyer EC (2009) Oxidative stress, lens gap junctions, and cataracts. Antioxid Redox Signal 11:339–353. doi:10.1089/ars.2008.2119
Bu L, Jin Y, Shi Y et al (2002) Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet 31:276–278. doi:10.1038/ng921
Cheng C, Ansari MM, Cooper JA, Gong X (2013) EphA2 and Src regulate equatorial cell morphogenesis during lens development. Development 140:4237–4245. doi:10.1242/dev.100727
Chorley BN, Campbell MR, Wang X et al (2012) Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha. Nucleic Acids Res 40:7416–7429. doi:10.1093/nar/gks409
Churchill A, Graw J (2011) Clinical and experimental advances in congenital and paediatric cataracts. Philos Trans R Soc Lond B Biol Sci 366:1234–1249. doi:10.1098/rstb.2010.0227
Congdon N, Broman KW, Lai H et al (2005) Cortical, but not posterior subcapsular, cataract shows significant familial aggregation in an older population after adjustment for possible shared environmental factors. Ophthalmology 112:73–77. doi:10.1016/j.ophtha.2004.07.012
Cvekl A, Ashery-Padan R (2014) The cellular and molecular mechanisms of vertebrate lens development. Development 141:4432–4447. doi:10.1242/dev.107953
Cvekl A, Duncan MK (2007) Genetic and epigenetic mechanisms of gene regulation during lens development. Prog Retin Eye Res 26:555–597. doi:10.1016/j.preteyeres.2007.07.002
Cvekl A, Sax CM, Bresnick EH, Piatigorsky J (1994) A complex array of positive and negative elements regulates the chicken alpha A-crystallin gene: involvement of Pax-6, USF, CREB and/or CREM, and AP-1 proteins. Mol Cell Biol 14:7363–7376
Donner AL, Episkopou V, Maas RL (2007) Sox2 and Pou2f1 interact to control lens and olfactory placode development. Dev Biol 303:784–799. doi:10.1016/j.ydbio.2006.10.047
Du P, Kibbe WA, Lin SM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548. doi:10.1093/bioinformatics/btn224
Fares-Taie L, Gerber S, Chassaing N et al (2013) ALDH1A3 mutations cause recessive anophthalmia and microphthalmia. Am J Hum Genet 92:265–270. doi:10.1016/j.ajhg.2012.12.003
Franceschini A, Szklarczyk D, Frankild S et al (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res 41:D808–D815. doi:10.1093/nar/gks1094
Fu L, Liang JJ-N (2002) Detection of protein–protein interactions among lens crystallins in a mammalian two-hybrid system assay. J Biol Chem 277:4255–4260. doi:10.1074/jbc.M110027200
Fujimoto M, Izu H, Seki K et al (2004) HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J 23:4297–4306. doi:10.1038/sj.emboj.7600435
Graw J, Klopp N, Neuhäuser-Klaus A et al (2002) Crygf(Rop): the first mutation in the Crygf gene causing a unique radial lens opacity. Invest Ophthalmol Vis Sci 43:2998–3002
Hammond CJ, Snieder H, Spector TD, Gilbert CE (2000) Genetic and environmental factors in age-related nuclear cataracts in monozygotic and dizygotic twins. N Engl J Med 342:1786–1790. doi:10.1056/NEJM200006153422404
Hammond CJ, Duncan DD, Snieder H et al (2001) The heritability of age-related cortical cataract: the twin eye study. Invest Ophthalmol Vis Sci 42:601–605
Hansen L, Eiberg H, Rosenberg T (2007) Novel MAF mutation in a family with congenital cataract–microcornea syndrome. Mol Vis 13:2019–2022
He M, Kratz LE, Michel JJ et al (2011) Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest 121:976–984. doi:10.1172/JCI42650
Hirotsu Y, Katsuoka F, Funayama R et al (2012) Nrf2–MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res 40:10228–10239. doi:10.1093/nar/gks827
Huang B, He W (2010) Molecular characteristics of inherited congenital cataracts. Eur J Med Genet 53:347–357. doi:10.1016/j.ejmg.2010.07.001
Huang DW, Sherman BT, Zheng X et al (2009) Extracting biological meaning from large gene lists with DAVID. Curr Protoc Bioinform. doi:10.1002/0471250953.bi1311s27 (Chapter 13:Unit 13.11)
Jamieson RV, Perveen R, Kerr B et al (2002) Domain disruption and mutation of the bZIP transcription factor, MAF, associated with cataract, ocular anterior segment dysgenesis and coloboma. Hum Mol Genet 11:33–42
Jolma A, Yan J, Whitington T et al (2013) DNA-binding specificities of human transcription factors. Cell 152:327–339. doi:10.1016/j.cell.2012.12.009
Kannan MB, Solovieva V, Blank V (2012) The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta 1823:1841–1846. doi:10.1016/j.bbamcr.2012.06.012
Kasaikina MV, Fomenko DE, Labunskyy VM et al (2011) Roles of the 15-kDa selenoprotein (Sep15) in redox homeostasis and cataract development revealed by the analysis of Sep 15 knockout mice. J Biol Chem 286:33203–33212. doi:10.1074/jbc.M111.259218
Kataoka K (2007) Multiple mechanisms and functions of maf transcription factors in the regulation of tissue-specific genes. J Biochem 141:775–781. doi:10.1093/jb/mvm105
Katsuoka F, Motohashi H, Tamagawa Y et al (2003) Small Maf compound mutants display central nervous system neuronal degeneration, aberrant transcription, and Bach protein mislocalization coincident with myoclonus and abnormal startle response. Mol Cell Biol 23:1163–1174
Kawauchi S, Takahashi S, Nakajima O et al (1999) Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem 274:19254–19260
Kim JI, Li T, Ho IC et al (1999) Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci 96:3781–3785
Lachke SA, Maas RL (2010) Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip Rev Syst Biol Med 2:305–323. doi:10.1002/wsbm.59
Lachke SA, Alkuraya FS, Kneeland SC et al (2011) Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 331:1571–1576. doi:10.1126/science.1195970
Lachke SA, Higgins AW, Inagaki M et al (2012a) The cell adhesion gene PVRL3 is associated with congenital ocular defects. Hum Genet 131:235–250. doi:10.1007/s00439-011-1064-z
Lachke SA, Ho JWK, Kryukov GV et al (2012b) iSyTE: integrated systems tool for eye gene discovery. Invest Ophthalmol Vis Sci 53:1617–1627. doi:10.1167/iovs.11-8839
Lassen N, Bateman JB, Estey T et al (2007) Multiple and additive functions of ALDH3A1 and ALDH1A1: cataract phenotype and ocular oxidative damage in Aldh3a1(−/−)/Aldh1a1(−/−) knock-out mice. J Biol Chem 282:25668–25676. doi:10.1074/jbc.M702076200
Li W, Yu S, Liu T et al (2008) Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim Biophys Acta 1783:1847–1856. doi:10.1016/j.bbamcr.2008.05.024
Litt M, Kramer P, LaMorticella DM et al (1998) Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet 7:471–474
Liu Y, Liu X, Zhang T et al (2007) Cytoprotective effects of proteasome beta5 subunit overexpression in lens epithelial cells. Mol Vis 13:31–38
Long Y, Li Q, Li J, Cui Z (2011) Molecular analysis, developmental function and heavy metal-induced expression of ABCC5 in zebrafish. Comp Biochem Physiol B Biochem Mol Biol 158:46–55. doi:10.1016/j.cbpb.2010.09.005
Maeda A, Moriguchi T, Hamada M et al (2009) Transcription factor GATA-3 is essential for lens development. Dev Dyn 238:2280–2291. doi:10.1002/dvdy.22035
Malhotra D, Portales-Casamar E, Singh A et al (2010) Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38:5718–5734. doi:10.1093/nar/gkq212
Min J-N, Zhang Y, Moskophidis D, Mivechi NF (2004) Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis 40:205–217. doi:10.1002/gene.20087
Mori M, Li G, Abe I et al (2006) Lanosterol synthase mutations cause cholesterol deficiency-associated cataracts in the Shumiya cataract rat. J Clin Invest 116:395–404. doi:10.1172/JCI20797
Motohashi H, O’Connor T, Katsuoka F et al (2002) Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1–12
Motohashi H, Katsuoka F, Engel JD, Yamamoto M (2004) Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1–Nrf2 regulatory pathway. Proc Natl Acad Sci 101:6379–6384. doi:10.1073/pnas.0305902101
Newburger DE, Bulyk ML (2009) UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res 37:D77–D82. doi:10.1093/nar/gkn660
Nishiguchi S, Wood H, Kondoh H et al (1998) Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev 12:776–781
Nongpiur ME, Khor CC, Jia H et al (2014) ABCC5, a gene that influences the anterior chamber depth, is associated with primary angle closure glaucoma. PLoS Genet 10:e1004089. doi:10.1371/journal.pgen.1004089
Ogino H, Yasuda K (1998) Induction of lens differentiation by activation of a bZIP transcription factor, L-Maf. Science 280:115–118
Onodera K, Shavit JA, Motohashi H et al (2000) Perinatal synthetic lethality and hematopoietic defects in compound mafG:mafK mutant mice. EMBO J 19:1335–1345. doi:10.1093/emboj/19.6.1335
Portales-Casamar E, Thongjuea S, Kwon AT et al (2010) JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res 38:D105–D110. doi:10.1093/nar/gkp950
Rao GN, Khanna R, Payal A (2011) The global burden of cataract. Curr Opin Ophthalmol 22:4–9. doi:10.1097/ICU.0b013e3283414fc8
Reed NA, Oh DJ, Czymmek KJ, Duncan MK (2001) An immunohistochemical method for the detection of proteins in the vertebrate lens. J Immunol Methods 253:243–252
Ring BZ, Cordes SP, Overbeek PA, Barsh GS (2000) Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127:307–317
Robasky K, Bulyk ML (2011) UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein–DNA interactions. Nucleic Acids Res 39:D124–D128. doi:10.1093/nar/gkq992
Rudolph T, Sjölander A, Palmér MS et al (2011) Ubiquitin carboxyl-terminal esterase LI (UCHLI) SI8Y polymorphism in patients with cataracts. Ophthalmic Genet 32:75–79. doi:10.3109/13816810.2010.544360
Scheiblin DA, Gao J, Caplan JL et al (2014) Beta-1 integrin is important for the structural maintenance and homeostasis of differentiating fiber cells. Int J Biochem Cell Biol 50:132–145. doi:10.1016/j.biocel.2014.02.021
Shaham O, Smith AN, Robinson ML et al (2009) Pax6 is essential for lens fiber cell differentiation. Development 136:2567–2578. doi:10.1242/dev.032888
Shavit JA, Motohashi H, Onodera K et al (1998) Impaired megakaryopoiesis and behavioral defects in mafG-null mutant mice. Genes Dev 12:2164–2174
Shiels A, Hejtmancik JF (2013) Genetics of human cataract. Clin Genet 84:120–127. doi:10.1111/cge.12182
Shiels A, King JM, Mackay DS, Bassnett S (2007) Refractive defects and cataracts in mice lacking lens intrinsic membrane protein-2. Invest Ophthalmol Vis Sci 48:500–508. doi:10.1167/iovs.06-0947
Shiels A, Bennett TM, Hejtmancik JF (2010) Cat-map: putting cataract on the map. Mol Vis 16:2007–2015
Sorokina EA, Muheisen S, Mlodik N, Semina EV (2011) MIP/Aquaporin 0 represents a direct transcriptional target of PITX3 in the developing lens. PLoS One 6:e21122. doi:10.1371/journal.pone.0021122
Stamenkovic M, Radic T, Stefanovic I et al (2014) Glutathione S-transferase omega-2 polymorphism Asn142Asp modifies the risk of age-related cataract in smokers and subjects exposed to ultraviolet irradiation. Clin Exp Ophthalmol 42:277–283. doi:10.1111/ceo.12180
Strzalka-Mrozik B, Prudlo L, Kimsa MW et al (2013) Quantitative analysis of SOD2, ALDH1A1 and MGST1 messenger ribonucleic acid in anterior lens epithelium of patients with pseudoexfoliation syndrome. Mol Vis 19:1341–1349
Tanaka T, Tsujimura T, Takeda K et al (1998) Targeted disruption of ATF4 discloses its essential role in the formation of eye lens fibres. Genes Cells 3:801–810
Toki T, Itoh J, Kitazawa J et al (1997) Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene 14:1901–1910. doi:10.1038/sj.onc.1201024
Vanita V, Singh D, Robinson PN et al (2006) A novel mutation in the DNA-binding domain of MAF at 16q23.1 associated with autosomal dominant “cerulean cataract” in an Indian family. Am J Med Genet A 140:558–566. doi:10.1002/ajmg.a.31126
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5:276–287. doi:10.1038/nrg1315
Wigle JT, Chowdhury K, Gruss P, Oliver G (1999) Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet 21:318–322. doi:10.1038/6844
Yahyavi M, Abouzeid H, Gawdat G et al (2013) ALDH1A3 loss of function causes bilateral anophthalmia/microphthalmia and hypoplasia of the optic nerve and optic chiasm. Hum Mol Genet 22:3250–3258. doi:10.1093/hmg/ddt179
Yamazaki H, Katsuoka F, Motohashi H et al (2012) Embryonic lethality and fetal liver apoptosis in mice lacking all three small Maf proteins. Mol Cell Biol 32:808–816. doi:10.1128/MCB.06543-11
Yang Y, Chauhan BK, Cveklova K, Cvekl A (2004) Transcriptional regulation of mouse alphaB- and gammaF-crystallin genes in lens: opposite promoter-specific interactions between Pax6 and large Maf transcription factors. J Mol Biol 344:351–368. doi:10.1016/j.jmb.2004.07.102
Yoshida T, Yasuda K (2002) Characterization of the chicken L-Maf, MafB and c-Maf in crystallin gene regulation and lens differentiation. Genes Cells 7:693–706
Zenkel M, Kruse FE, Naumann GOH, Schlötzer-Schrehardt U (2007) Impaired cytoprotective mechanisms in eyes with pseudoexfoliation syndrome/glaucoma. Invest Ophthalmol Vis Sci 48:5558–5566. doi:10.1167/iovs.07-0750
Acknowledgments
We thank Drs. Melinda Duncan and Irfan Saadi for helpful comments and discussions. This work was supported by National Institutes of Health [R01 EY021505 to S.L.]; and Fight for Sight Grant-in-Aid award to S.L. S.L. is a Pew Scholar in Biomedical Sciences.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Additional information
S. A. Agrawal, D. Anand and A. D. Siddam contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Agrawal, S.A., Anand, D., Siddam, A.D. et al. Compound mouse mutants of bZIP transcription factors Mafg and Mafk reveal a regulatory network of non-crystallin genes associated with cataract. Hum Genet 134, 717–735 (2015). https://doi.org/10.1007/s00439-015-1554-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-015-1554-5