Abstract
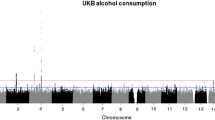
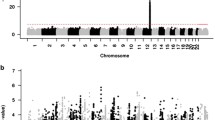
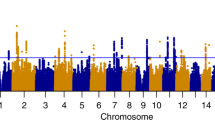
Maximum number of alcoholic drinks consumed in a 24-h period (maxdrinks) is a heritable (>50 %) trait and is strongly correlated with vulnerability to excessive alcohol consumption and subsequent alcohol dependence (AD). Several genome-wide association studies (GWAS) have studied alcohol dependence, but few have concentrated on excessive alcohol consumption. We performed two GWAS using maxdrinks as an excessive alcohol consumption phenotype: one in 118 extended families (N = 2,322) selected from the Collaborative Study on the Genetics of Alcoholism (COGA), and the other in a case–control sample (N = 2,593) derived from the Study of Addiction: Genes and Environment (SAGE). The strongest association in the COGA families was detected with rs9523562 (p = 2.1 × 10−6) located in an intergenic region on chromosome 13q31.1; the strongest association in the SAGE dataset was with rs67666182 (p = 7.1 × 10−7), located in an intergenic region on chromosome 8. We also performed a meta-analysis with these two GWAS and demonstrated evidence of association in both datasets for the LMO1 (p = 7.2 × 10−7) and PLCL1 genes (p = 4.1 × 10−6) with maxdrinks. A variant in AUTS2 and variants in INADL, C15orf32 and HIP1 that were associated with measures of alcohol consumption in a meta-analysis of GWAS studies and a GWAS of alcohol consumption factor score also showed nominal association in the current meta-analysis. The present study has identified several loci that warrant further examination in independent samples. Among the top SNPs in each of the dataset (p ≤ 10−4) far more showed the same direction of effect in the other dataset than would be expected by chance (p = 2 × 10−3, 3 × 10−6), suggesting that there are true signals among these top SNPs, even though no SNP reached genome-wide levels of significance.

Similar content being viewed by others
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. doi:10.1038/nmeth0410-248
Agrawal A, Freedman ND, Cheng YC, Lin P, Shaffer JR, Sun Q, Taylor K, Yaspan B, Cole JW, Cornelis MC, DeSensi RS, Fitzpatrick A, Heiss G, Kang JH, O’Connell J, Bennett S, Bookman E, Bucholz KK, Caporaso N, Crout R, Dick DM, Edenberg HJ, Goate A, Hesselbrock V, Kittner S, Kramer J, Nurnberger JI Jr, Qi L, Rice JP, Schuckit M, van Dam RM, Boerwinkle E, Hu F, Levy S, Marazita M, Mitchell BD, Pasquale LR, Bierut LJ (2012) Measuring alcohol consumption for genomic meta-analyses of alcohol intake: opportunities and challenges. Am J Clin Nutr 95(3):539–547. doi:10.3945/ajcn.111.015545
Allen NE, Beral V, Casabonne D, Kan SW, Reeves GK, Brown A, Green J (2009) Moderate alcohol intake and cancer incidence in women. J Natl Cancer Inst 101(5):296–305. doi:10.1093/jnci/djn514
Baik I, Cho NH, Kim SH, Han BG, Shin C (2011) Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr 93(4):809–816. doi:10.3945/ajcn.110.001776
Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP (2010) A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA 107(11):5082–5087. doi:10.1073/pnas.0911109107
Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ (2012) ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry 17(4):445–450. doi:10.1038/mp.2011.124
Brooks PJ, Zakhari S (2013) Moderate alcohol consumption and breast cancer in women: from epidemiology to mechanisms and interventions. Alcohol Clin Exp Res 37(1):23–30. doi:10.1111/j.1530-0277.2012.01888.x
Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81(5):1084–1097. doi:10.1086/521987
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI Jr, Reich T, Schmidt I, Schuckit MA (1994) A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 55(2):149–158
Caan W, de Belleroche J (2002) Drink, drugs and dependence: from science to clinical practice. Taylor & Francis Group, UK
Chen MH, Yang Q (2010) GWAF: an R package for genome-wide association analyses with family data. Bioinformatics 26(4):580–581. doi:10.1093/bioinformatics/btp710
Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ (1999) Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet 65(3):795–807. doi:10.1086/302540
Chen WY, Rosner B, Hankinson SE, Colditz GA, Willett WC (2011) Moderate alcohol consumption during adult life, drinking patterns, and breast cancer risk. JAMA J Am Med Assoc 306(17):1884–1890. doi:10.1001/jama.2011.1590
Dudley KJ, Li X, Kobor MS, Kippin TE, Bredy TW (2011) Epigenetic mechanisms mediating vulnerability and resilience to psychiatric disorders. Neurosci Biobehav Rev 35(7):1544–1551. doi:10.1016/j.neubiorev.2010.12.016
Edenberg HJ (2007) The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health J Natl Inst Alcohol Abuse Alcohol 30(1):5–13
Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI Jr, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T (2010) Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res 34(5):840–852. doi:10.1111/j.1530-0277.2010.01156.x
Grant JD, Agrawal A, Bucholz KK, Madden PA, Pergadia ML, Nelson EC, Lynskey MT, Todd RD, Todorov AA, Hansell NK, Whitfield JB, Martin NG, Heath AC (2009) Alcohol consumption indices of genetic risk for alcohol dependence. Biol Psychiatry 66(8):795–800. doi:10.1016/j.biopsych.2009.05.018
Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW (2011) A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol Psychiatry 70(6):513–518. doi:10.1016/j.biopsych.2011.02.028
Hurley TD, Edenberg HJ (2012) Genes encoding enzymes involved in ethanol metabolism. Alcohol Res Curr Rev 34(3):339–344
Jiang H, Wu D, Simon MI (1994) Activation of phospholipase C beta 4 by heterotrimeric GTP-binding proteins. J Biol Chem 269(10):7593–7596
Kendler KS, Myers J, Dick D, Prescott CA (2010) The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol Clin Exp Res 34(6):1058–1065. doi:10.1111/j.1530-0277.2010.01181.x
Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV (2011) Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol Clin Exp Res 35(5):963–975. doi:10.1111/j.1530-0277.2010.01427.x
Lasek AW, Giorgetti F, Berger KH, Tayor S, Heberlein U (2011) Lmo genes regulate behavioral responses to ethanol in Drosophila melanogaster and the mouse. Alcohol Clin Exp Res 35(9):1600–1606. doi:10.1111/j.1530-0277.2011.01506.x
Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, Macgregor S (2010) A versatile gene-based test for genome-wide association studies. Am J Hum Genet 87(1):139–145. doi:10.1016/j.ajhg.2010.06.009
Mattick JS (2009) The genetic signatures of noncoding RNAs. PLoS Genet 5(4):e1000459. doi:10.1371/journal.pgen.1000459
Mokdad AH, Marks JS, Stroup DF, Gerberding JL (2004) Actual causes of death in the United States, 2000. JAMA J Am Med Assoc 291(10):1238–1245. doi:10.1001/jama.291.10.1238
Muller D, Koch RD, von Specht H, Volker W, Munch EM (1985) Neurophysiologic findings in chronic alcohol abuse. Psychiatrie Neurol Med Psychol 37(3):129–132
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904–909. doi:10.1038/ng1847
Purcell S, Cherny SS, Sham PC (2003) Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19(1):149–150
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. doi:10.1086/519795
Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP (2000) A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet 96(5):632–637
Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P (2011) Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci USA 108(17):7119–7124. doi:10.1073/pnas.1017288108
Testino G (2008) Alcoholic diseases in hepato-gastroenterology: a point of view. Hepatogastroenterology 55(82–83):371–377
Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet 48(4):677–681
Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M (2009) Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 66(7):773–784. doi:10.1001/archgenpsychiatry.2009.83
Wang K, Diskin SJ, Zhang H, Attiyeh EF, Winter C, Hou C, Schnepp RW, Diamond M, Bosse K, Mayes PA, Glessner J, Kim C, Frackelton E, Garris M, Wang Q, Glaberson W, Chiavacci R, Nguyen L, Jagannathan J, Saeki N, Sasaki H, Grant SF, Iolascon A, Mosse YP, Cole KA, Li H, Devoto M, McGrady PW, London WB, Capasso M, Rahman N, Hakonarson H, Maris JM (2011) Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature 469(7329):216–220. doi:10.1038/nature09609
Wang JC, Foroud T, Hinrichs AL, Le NX, Bertelsen S, Budde JP, Harari O, Koller DL, Wetherill L, Agrawal A, Almasy L, Brooks AI, Bucholz K, Dick D, Hesselbrock V, Johnson EO, Kang S, Kapoor M, Kramer J, Kuperman S, Madden PA, Manz N, Martin NG, McClintick JN, Montgomery GW, Nurnberger JI Jr, Rangaswamy M, Rice J, Schuckit M, Tischfield JA, Whitfield JB, Xuei X, Porjesz B, Heath AC, Edenberg HJ, Bierut LJ, Goate AM (2012a) A genome-wide association study of alcohol-dependence symptom counts in extended pedigrees identifies C15orf53. Mol Psychiatry. doi:10.1038/mp.2012.143mp2012143
Wang JC, Kapoor M, Goate AM (2012b) The genetics of substance dependence. Annu Rev Genomics Hum Genet 13:241–261. doi:10.1146/annurev-genom-090711-163844
Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40((Database issue)):D930–D934. doi:10.1093/nar/gkr917
Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26(17):2190–2191. doi:10.1093/bioinformatics/btq340
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82. doi:10.1016/j.ajhg.2010.11.011
Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, Eleftheriadis M, Sinning CR, Schnabel RB, Lubos E, Mennerich D, Rust W, Perret C, Proust C, Nicaud V, Loscalzo J, Hubner N, Tregouet D, Munzel T, Ziegler A, Tiret L, Blankenberg S, Cambien F (2010) Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS ONE 5(5):e10693. doi:10.1371/journal.pone.0010693
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA): COGA, Principal Investigators B Porjesz, V Hesselbrock, H Edenberg, L Bierut includes ten different centers: University of Connecticut (V Hesselbrock); Indiana University (HJ Edenberg, J Nurnberger Jr, T Foroud); University of Iowa (S Kuperman, J Kramer); SUNY Downstate (B Porjesz); Washington University in Saint Louis (L Bierut, A Goate, J Rice, K Bucholz); University of California at San Diego (M Schuckit); Rutgers University (J Tischfield); Southwest Foundation (L Almasy), Howard University (R Taylor) and Virginia Commonwealth University (D Dick). A Parsian and M Reilly are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P Michael Conneally, Raymond Crowe and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
The Study of Addiction: Genetics and Environment (SAGE): Funding support for SAGE was provided through the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004422). SAGE is one of the GWAS funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of data sets and samples was provided by COGA (U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392) and the Family Study of Cocaine Dependence (FSCD; R01 DA013423, R01 DA019963). Genotyping at the Johns Hopkins University Center for Inherited Disease Research was supported by the NIH GEI (U01HG004438) Grant, NIAAA, NIDA and the NIH contract ‘High throughput genotyping for studying the genetic contributions to human disease’.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kapoor, M., Wang, JC., Wetherill, L. et al. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum Genet 132, 1141–1151 (2013). https://doi.org/10.1007/s00439-013-1318-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-013-1318-z