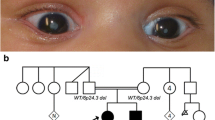
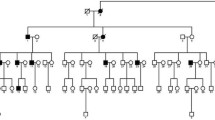
Abstract
We performed homozygosity mapping in a consanguineous Pakistani family segregating autosomal-recessive congenital cataracts and identified linkage to a 3.03 Mb locus on chromosome 6p24 containing the GCNT2 gene. GCNT2 encodes glucosaminyl (N-acetyl) transferase 2, an enzyme responsible for the formation of the blood group I antigen. Rare biallelic GCNT2 mutations have been shown to cause the association of congenital cataracts and the adult i blood group, making GCNT2 the prime candidate gene for the observed phenotype. Indeed, we identified a homozygous deletion segregating with cataracts that encompasses exons 1B, 1C, 2 and 3 of GCNT2. Long-range polymerase chain reaction and breakpoint sequencing revealed that affected individuals in this and in a second, apparently unrelated Pakistani family segregating congenital cataracts are homozygous for the same 93 kb deletion. The deletion is flanked by Alu repeats of the AluS family on both sides and microsatellite genotyping suggested that its occurrence in the two families was the product of recurrent Alu–Alu repeat-mediated nonhomologous recombinations or an old founder effect. Subsequently, we showed that cataract-affected individuals in both families have the adult i blood group, whereas unaffected individuals have blood group I as the vast majority of the population. Because the GCNT2 locus is rich in Short INterspersed Elements (SINE repeats) and thus likely prone to genomic rearrangements, microdeletions or microduplications at this locus might cause a larger than currently anticipated fraction of apparently isolated autosomal-recessive cataracts.



Similar content being viewed by others
References
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743
Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101
Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379
Chaplin H, Hunter VL, Malecek AC, Kilzer P, Rosche ME (1986) Clinically significant allo-anti-I in an I-negative patient with massive hemorrhage. Transfusion 26:57–61
Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, Fitzgerald T, Hu M, Ihm CH, Kristiansson K, Macarthur DG, Macdonald JR, Onyiah I, Pang AW, Robson S, Stirrups K, Valsesia A, Walter K, Wei J, Tyler-Smith C, Carter NP, Lee C, Scherer SW, Hurles ME (2010) Origins and functional impact of copy number variation in the human genome. Nature 464:704–712
Deininger PL, Batzer MA (1999) Alu repeats and human disease. Mol Genet Metab 67:183–193
Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, Diekhans M, Dreszer TR, Giardine BM, Harte RA, Hillman-Jackson J, Hsu F, Kirkup V, Kuhn RM, Learned K, Li CH, Meyer LR, Pohl A, Raney BJ, Rosenbloom KR, Smith KE, Haussler D, Kent WJ (2011) The UCSC genome browser database: update 2011. Nucleic Acids Res 39:D876–D882
Garshasbi M, Motazacker MM, Kahrizi K, Behjati F, Abedini SS, Nieh SE, Firouzabadi SG, Becker C, Ruschendorf F, Nurnberg P, Tzschach A, Vazifehmand R, Erdogan F, Ullmann R, Lenzner S, Kuss AW, Ropers HH, Najmabadi H (2006) SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum Genet 118:708–715
Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13
Hejtmancik JF (2008) Congenital cataracts and their molecular genetics. Semin Cell Dev Biol 19:134–149
Inaba N, Hiruma T, Togayachi A, Iwasaki H, Wang XH, Furukawa Y, Sumi R, Kudo T, Fujimura K, Iwai T, Gotoh M, Nakamura M, Narimatsu H (2003) A novel I-branching beta-1, 6-N-acetylglucosaminyltransferase involved in human blood group I antigen expression. Blood 101:2870–2876
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Li W, Freudenberg J (2009) Two-parameter characterization of chromosome-scale recombination rate. Genome Res 19:2300–2307
Lin M, Hou MJ, Yu LC (2006) A novel IGnT allele responsible for the adult i phenotype. Transfusion 46:1982–1987
Liquori CL, Berg MJ, Squitieri F, Leedom TP, Ptacek L, Johnson EW, Marchuk DA (2007) Deletions in CCM2 are a common cause of cerebral cavernous malformations. Am J Hum Genet 80:69–75
Lupski JR, Stankiewicz P (2005) Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1:e49
O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266
Pras E, Raz J, Yahalom V, Frydman M, Garzozi HJ, Pras E, Hejtmancik JF (2004) A nonsense mutation in the glucosaminyl (N-acetyl) transferase 2 gene (GCNT2): association with autosomal recessive congenital cataracts. Invest Ophthalmol Vis Sci 45:1940–1945
Ringpfeil F, Nakano A, Uitto J, Pulkkinen L (2001) Compound heterozygosity for a recurrent 16.5-kb Alu-mediated deletion mutation and single-base-pair substitutions in the ABCC6 gene results in pseudoxanthoma elasticum. Am J Hum Genet 68:642–652
Ruschendorf F, Nurnberg P (2005) ALOHOMORA: a tool for linkage analysis using 10K SNP array data. Bioinformatics 21:2123–2125
Sajjad N, Goebel I, Kakar N, Cheema AM, Kubisch C, Ahmad J (2008) A novel HSF4 gene mutation (p.R405X) causing autosomal recessive congenital cataracts in a large consanguineous family from Pakistan. BMC Med Genet 9:99
Shiels A, Bennett TM, Hejtmancik JF (2010) Cat-Map: putting cataract on the map. Mol Vis 16:2007–2015
van der Zwaag B, Franke L, Poot M, Hochstenbach R, Spierenburg HA, Vorstman JA, van Daalen E, de Jonge MV, Verbeek NE, Brilstra EH, van ‘t Slot R, Ophoff RA, van Es MA, Blauw HM, Veldink JH, Buizer-Voskamp JE, Beemer FA, van den Berg LH, Wijmenga C, van Amstel HK, van Engeland H, Burbach JP, Staal WG et al (2009) Gene-network analysis identifies susceptibility genes related to glycobiology in autism. PLoS One 4:e5324
Yu LC, Twu YC, Chang CY, Lin M (2001) Molecular basis of the adult i phenotype and the gene responsible for the expression of the human blood group I antigen. Blood 98:3840–3845
Yu LC, Twu YC, Chou ML, Reid ME, Gray AR, Moulds JM, Chang CY, Lin M (2003) The molecular genetics of the human I locus and molecular background explain the partial association of the adult i phenotype with congenital cataracts. Blood 101:2081–2088
Acknowledgments
We wish to thank the family members for their cooperation and participation in this study. G.B. was supported by the Deutsche Forschungsgemeinschaft (DFG; BO2985/3-1). This study was supported by the generous help of Ahmed Farooq Bazai, Vice Chancellor, BUITEMS, through faculty funds. We thank Dr. Sigrid Roters (Cologne, Germany) for ophthalmologic advice.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
439_2011_1062_MOESM2_ESM.tif
Supplementary Figure 1 Linkage of autosomal-recessive cataracts to chromosome 6p24 in family CA03. Schematic representation of genome-wide LOD score calculations in family CA03. LOD scores calculated with ALLEGRO are given along the y-axis relative to genomic positionin centi Morgan (cM) on the x-axis. Note a single significant peak (LOD score 3.01) located on chromosome 6. (DOC 1.45 MB)
439_2011_1062_MOESM3_ESM.tif
Supplementary Figure 2 Enrichment of the GCTN2 region on 6p24 for SINE repeats.Enrichment of SINE repeats in shifting windows of size 100kb on chromosome 6 (a) and around GCNT2 (b). See methods for details on the calculation of the enrichment score. (TIF 1.63 MB)
439_2011_1062_MOESM4_ESM.tif
Supplementary Figure 3 Individuals homozygous for the GCNT2 deletion have the adult i blood group.a Test tube results of anti-I typing in three members of families CA03 and CA05 each, showing that individuals affected by cataracts and homozygous for the GCNT2 deletion have the adult i blood group (I-negative) whereas unaffected family members have the common I blood group.b Test tube results of anti-I typing for two unrelated healthy Pakistani controls (control 1 and control 2) as well as a negative agglutination control (cord blood, I negative, i positive) and a positive agglutination control (adult donor blood, I positive, i negative). (TIF 3.11 MB)
Rights and permissions
About this article
Cite this article
Borck, G., Kakar, N., Hoch, J. et al. An Alu repeat-mediated genomic GCNT2 deletion underlies congenital cataracts and adult i blood group. Hum Genet 131, 209–216 (2012). https://doi.org/10.1007/s00439-011-1062-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-011-1062-1