Abstract
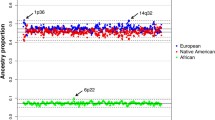
Migrations to the new world brought together individuals from Europe, Africa and the Americans. Inter-mating between these migrant and indigenous populations led to the subsequent formation of new admixed populations, such as African and Latino Americans. These unprecedented events brought together genomes that had evolved independently on different continents for tens of thousands of years and presented new environmental challenges for the indigenous and migrant populations, as well as their offspring. These circumstances provided novel opportunities for natural selection to occur that could be reflected in deviations at specific locations from the genome-wide ancestry distribution. Here we present an analysis examining European, Native American and African ancestry based on 284 microsatellite markers in a study of Mexican Americans from the Family Blood Pressure Program. We identified two genomic regions where there was a significant decrement in African ancestry (at 2p25.1, p < 10−8 and 9p24.1, p < 2 × 10−5) and one region with a significant increase in European ancestry (at 1p33, p < 2 × 10−5). These locations may harbor genes that have been subjected to natural selection after the ancestral mixing giving rise to Mexicans.




Similar content being viewed by others
References
Bonilla C, Gutierrez G, Parra EJ, Kline C, Shriver MD (2005) Admixture analysis of a rural population of the state of Guerrero, Mexico. Am J Phys Anthropol 128:861–869
Chakraborty R, Weiss KM (1986) Frequencies of complex diseases in hybrid populations. Am J Phys Anthrop 70:489–503
Chakraborty R, Weiss KM (1988) Admixture as a tool for finding linked genes and detecting that difference from allelic association between loci. PNAS 85:9119–9123
Collins-Schramm HE, Phillips CM, Operario DJ, Lee JS, Weber JL, Hanson RL, Knowler WC, Cooper R, Li H, Seldin MF (2002) Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am J Hum Genet 70:737–750
Crow JF, Kimura M (1970) An introduction to population genetics theory. Harper, Row
Daniels PR, Kardia SL, Hanis CL, Brown CA, Hutchinson R, Boerwinkle E, Turner ST, Genetic Epidemiology Network of Arteriopathy study (2004) Familial aggregation of hypertension treatment and control in the Genetic Epidemiology Network of Arteriopathy (GENOA) study. Am J Med 116:676–681
Efron B, Tibshirani R (1993) An Introduction to the Bootstrap. Chapman & Hall, London
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
FBPP Investigators (2002) Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP). Hypertension 39:3–9
Fernandez JR, Shriver MD, Beasley TM, Rafla-Demetrious N, Parra E, Albu J, Nicklas B, Ryan AS, McKeigue PM, Hoggart CL, Weinsier RL, Allison DB (2003) Association of African genetic admixture with resting metabolic rate and obesity among women. Obes Res 11:904–911
Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ (1991) Origins of US Hispanics Implications for diabetes. Diab care: 618–627
Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM (2004) Design and analysis of admixture mapping studies. Am J Hum Genet 74:965–978
Kimura M (1955) Solution of a process of random genetic drift with a continuous model. PNAS 41:144–150
Long JC (1991) The genetic structure of admixed populations. Genetics 127:417–428
McKeigue PM (1998) Mapping genes that underlie ethnic differences in disease risk: methods for detecting linkage in admixed populations, by conditioning on parental admixture. Am J Hum Genet 63:241–251
Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D (2004) Methods for high-density admixture mapping of disease genes. Am J Hum Genet 74:979–1000
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Reed TE (1969) Caucasian genes in American Negroes. Science 165:762–768
Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, DeLoa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BA, Hauser SL, Oksenberg JR, Hafler DA (2005) A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nat genet 37:1113–1118
Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, Choy E, Hu D, Tamraz B, Pawlikowska L, Wassel-Fyr C, Huntsman S, Waliszewska A, Rossin E, Li R, Garcia M, Reiner A, Ferrell R, Cummings S, Kwok PY, Harris T, Zmuda JM, Ziv E, Aging Health Body Composition (Health ABC) Study (2007) Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am J Hum Genet 80:716–726
Risch N (1992) Mapping genes for complex diseases using association studies in admixed populations. Am J Hum Genet 51:13
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA (2003) Skin pigmentation, biogeographical ancestry and admixture mapping. Hum genet 112:387–399
Smith MW, Lautenberger JA, Shin HD, Chretien JP, Shrestha S, Gilbert DA, O’Brien SJ (2001) Markers for mapping by admixture linkage disequilibrium in African American and Hispanic populations. Am J Hum Genet 69:1080–1094
Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E, De Jager PL, Mignault AA, Yi Z, De The G, Essex M, Sankale JL, Moore JH, Poku K, Phair JP, Goedert JJ, Vlahov D, Williams SM, Tishkoff SA, Winkler CA, De La Vega FM, Woodage T, Sninsky JJ, Hafler DA, Altshuler D, Gilbert DA, O’Brien SJ, Reich D (2004) A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet 74:1001–1013
Stephens JC, Briscoe D, O’Brien SJ (1994) Mapping by admixture linkage disequilibrium in human populations: limits and guidelines. Am J Hum Genet 55:809–824
Tang H, Peng J, Wang P, Risch NJ (2005) Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol 28:289–301
Tang H, Jorgenson E, Gadde M, Kardia SL, Rao DC, Zhu X, Schork NJ, Hanis CL, Risch N (2006) Racial admixture and its impact on BMI and blood pressure in African and Mexican Americans. Hum Genet 119:624–633
Tang H, Choudhry S, Mei R, Morgan M, Rodriguez-Cintron W, Burchard EG, Risch NJ (2007) Recent genetic selection in the ancestral admixture of Puerto Ricans. Am J Hum Genet 81:626–633
Wang S, Ray N, Rojas W, Parra MV, Bedoya G et al (2008) Geographic patterns of genome admixture in Latin American Mestizos. PLoS Genet 4(3):e10000037. doi:10.1371/journal.pgen.1000037
Zhu X, Cooper RS, Elston RC (2004) Linkage analysis of a complex disease through use of admixed populations. Am J Hum Genet 74:1136–1153
Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, Gu CC, Tang H, Rao DC, Risch N, Weder A (2005) Admixture mapping for hypertension loci with genome-scan markers. Nat Genet 37:177–181
Acknowledgments
This work was supported by grants awarded to the Family Blood Pressure Program, which is supported by a series of cooperative agreements from the National Heart, Lung and Blood Institute to GenNet, HyperGEN, GENOA and SAPPHIRe.
Conflict of interest statement
The authors declare that they have no competing financial interests
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
GenNet Network: Alan B. Weder (Network Director), Lillian Gleiberman (Network Coordinator), Anne E. Kwitek, Aravinda Chakravarti, Richard S. Cooper, Carolina Delgado, Howard J. Jacob, and Nicholas J. Schork
GENOA Network: Eric Boerwinkle (Network Director), Tom Mosley, Alanna Morrison, Kathy Klos, Craig Hanis, Sharon Kardia, and Stephen Turner
HyperGEN Network: Steven C. Hunt (Network Director), Janet Hood, Donna Arnett, John H. Eckfeldt, R. Curtis Ellison, Chi Gu, Gerardo Heiss, Paul Hopkins, Aldi T. Kraja, Jean-Marc Lalouel, Mark Leppert, Albert Oberman, Michael A. Province, D.C. Rao, Treva Rice, and Robert Weiss
SAPPHIRe Network: David Curb (Network Director), David Cox, Timothy Donlon, Victor Dzau, John Grove, Kamal Masaki, Richard Myers, Richard Olshen, Richard Pratt, Tom Quertermous, Neil Risch and Beatriz Rodriguez
National Heart, Lung, and Blood Institute: Dina Paltoo and Cashell E. Jaquish. Website: http://www.biostat.wustl.edu/fbpp/FBPP.shtml
Rights and permissions
About this article
Cite this article
Basu, A., Tang, H., Zhu, X. et al. Genome-wide distribution of ancestry in Mexican Americans. Hum Genet 124, 207–214 (2008). https://doi.org/10.1007/s00439-008-0541-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-008-0541-5