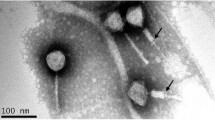
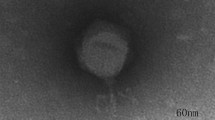
Abstract
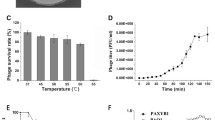
Bacteriophages are potential antibiotic substitutes for the treatment of antibiotic resistant bacteria. Here, we report the genome sequences of a double-stranded DNA podovirus vB_Pae_HB2107-3I against clinical multi-drug resistant Pseudomonas aeruginosa. Phage vB_Pae_HB2107-3I remained stable over a wide range of temperatures (37–60 °C) and pH values (pH 4–12). At MOI of 0.01, the latent period of vB_Pae_HB2107-3I was 10 min, and the final titer reached about 8.1 × 109 PFU/mL. The vB_Pae_HB2107-3I genome is 45,929 bp, with an average G + C content of 57%. A total of 72 open reading frames (ORFs) were predicted, of which 22 ORFs have a predicted function. Genome analyses confirmed the lysogenic nature of this phage. Phylogenetic analysis revealed that phage vB_Pae_HB2107-3I was a novel member of Caudovirales infecting P. aeruginosa. The characterization of vB_Pae_HB2107-3I enrich the research on Pseudomonas phages and provide a promising biocontrol agent against P. aeruginosa infections.





Similar content being viewed by others
Data availability
The complete genome sequence of Pseudomonas phage vB_Pae_HB2107-3I was deposited in the GenBank database under the accession number ON778006.
References
Amgarten D, Martins LF, Lombardi KC, Antunes LP, Silva de Souza AP, Nicastro GG, Kitajima EW, Quaggio RB, Upton C, Setubal JC, da Silva AM (2017) Three novel Pseudomonas phages isolated from composting provide insights into the evolution and diversity of tailed phages. BMC Genom 18:346
Cochran PK, Paul JH (1998) Seasonal abundance of lysogenic bacteria in a subtropical estuary. Appl Environ Microb 64:2308–2312
Curran CS, Bolig T, Torabi-Parizi P (2018) Mechanisms and targeted therapies for Pseudomonas aeruginosa lung infection. Am J Respir Crit Care Med 197:708–727
Dedrick RM, Jacobs-Sera D, Bustamante CAG, Garlena RA, Mavrich TN, Pope WH, Reyes JCC, Russell DA, Adair T, Alvey R, Bonilla JA, Bricker JS, Brown BR, Byrnes D, Cresawn SG, Davis WB, Dickson LA, Edgington NP, Findley AM, Golebiewska U, Grose JH, Hayes CF, Hughes LE, Hutchison KW, Isern S, Johnson AA, Kenna MA, Klyczek KK, Mageeney CM, Michael SF, Molloy SD, Montgomery MT, Neitzel J, Page ST, Pizzorno MC, Poxleitner MK, Rinehart CA, Robinson CJ, Rubin MR, Teyim JN, Vazquez E, Ware VC, Washington J, Hatfull GF (2017) Prophage-mediated defence against viral attack and viral counter-defence. Nat Microbiol 2:16251
Ding T, Sun H, Pan Q, Zhao F, Zhang Z, Ren H (2020) Isolation and characterization of vibrio parahaemolyticus bacteriophage vB_VpaS_PG07. Virus Res 286:198080
Edgar R, Friedman N, Molshanski-Mor S, Qimron U (2012) Reversing bacterial resistance to antibiotics by phage-mediated delivery of dominant sensitive genes. Appl Environ Microbiol 78:744–751
Ghosh D, Roy K, Williamson KE, White DC, Wommack KE, Sublette KL, Radosevich M (2008) Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl Environ Microb 74:495–502
Golkar Z, Bagasra O, Pace DG (2014) Bacteriophage therapy: a potential solution for the antibiotic resistance crisis. J Infect Dev Ctries 8:129–136
Gordillo Altamirano Fernando L, Barr Jeremy J (2019) Phage therapy in the Postantibiotic Era. Clin Microbiol Rev 32:e00066-e118
Harrison E, Brockhurst MA (2017) Ecological and evolutionary benefits of temperate phage: what does or doesn’t kill you makes you stronger. BioEssays 39:1700112
Joensen KG, Scheutz F, Lund O, Hasman H, Kaas RS, Nielsen EM, Aarestrup FM (2014) Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of Verotoxigenic Escherichia coli. J Clin Microbiol 52:1501–1510
Kilcher S, Studer P, Muessner C, Klumpp J, Loessner MJ (2018) Cross-genus rebooting of custom-made, synthetic bacteriophage genomes in L-form bacteria. Proc Natl Acad Sci USA 115:567–572
Kobiler O, Rokney A, Friedman N, Court DL, Stavans J, Oppenheim AB (2005) Quantitative kinetic analysis of the bacteriophage lambda genetic network. Proc Natl Acad Sci USA 102:4470–4475
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Latino L, Essoh C, Blouin Y, Hoang VuT, Pourcel C (2014) A novel Pseudomonas aeruginosa bacteriophage, Ab31, a chimera formed from temperate phage PAJU2 and P. Putida lytic phage AF: characteristics and mechanism of bacterial resistance. Plos One 9:e93777
Lu Z, Breidt F, Fleming HP, Altermann E, Klaenhammer TR (2003) Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int J Food Microbiol 84:225–235
McDaniel L, Paul JH (2005) Effect of nutrient addition and environmental factors on prophage induction in natural populations of Marine Synechococcus spe cies. Appl Environ Microb 71:842–850
Monteiro R, Pires DP, Costa AR, Azeredo J (2019) Phage therapy: going temperate? Trends Microbiol 27:368–378
Nowicki G, Walkowiak-Nowicka K, Zemleduch-Barylska A, Mleczko A, Frackowiak P, Nowaczyk N, Kozdrowska E, Barylski J (2017) Complete genome sequences of two novel autographiviruses infecting a bacterium from the Pseudomonas fluorescens group. Adv Virol 162:2907–2911
Park JY, Moon BY, Park JW, Thornton JA, Park YH, Seo KS (2017) Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci Rep 7:44929
Pires DP, Boas DV, Sillankorva S, Azeredo J (2015) Phage therapy: a step forward in the treatment of Pseudomonas aeruginosa infections. J Virol 89:7449–7456
Pirnay J-P, De Vos D, Verbeken G, Merabishvili M, Chanishvili N, Vaneechoutte M, Zizi M, Laire G, Lavigne R, Huys I, Van den Mooter G, Buckling A, Debarbieux L, Pouillot F, Azeredo J, Kutter E, Dublanchet A, Gorski A, Adamia R (2011) The phage therapy paradigm: prt-a -porter or sur-mesure? Pharm Res 28:934–937
Poole K (2011) Pseudomonas aeruginosa: resistance to the max. Front Microbiol 2:65
Qiao X, Sun Y, Qiao J, Di Sanzo F, Mindich L (2010) Characterization of Phi 2954, a newly isolated bacteriophage containing three dsRNA genomic segments. BMC Microbiol 10:55
Rao VB, Feiss M (2008) The bacteriophage DNA packaging motor. Annu Rev Genet 42:647–681
Roach DR, Debarbieux L (2017) Phage therapy: awakening a sleeping giant. Emerg Topics Life Sci 1:93–103
Shahin K, Bouzari M (2018) Bacteriophage application for biocontrolling Shigella flexneri in contaminated foods. J Food Sci Technol-Mysore 55:550–559
Thomas JA, Rolando MR, Carroll CA, Shen PS, Belnap DM, Weintraub ST, Serwer P, Hardies SC (2008) Characterization of Pseudomonas chlororaphis myovirus 201 phi 2–1 via genomic sequencing, mass spectrometry, and electron microscopy. Virology 376:330–338
Touchon M, Bernheim A, Rocha EPC (2016) Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J 10:2744–2754
Uchiyama J, Rashel M, Matsumoto T, Sumiyama Y, Wakiguchi H, Matsuzaki S (2009) Characteristics of a novel Pseudomonas aeruginosa bacteriophage, PAJU2, which is genetically related to bacteriophage D3. Virus Res 139:131–134
Wang R, Xing S, Zhao F, Li P, Mi Z, Shi T et al (2018) Characterization and genome analysis of novel phage vB_EfaP_IME195 infecting Enterococcus faecalis. Virus Genes 54:804–811
Williamson SJ, Houchin LA, Mcdaniel L, Paul JH (2002) Seasonal variation in lysogeny as depicted by prophage induction in Tampa Bay, Florida. Appl Environ Microb 68:4307–4314
Wu X, Wu Y, Tang Y, Gan B (2018) Genomic characterization and phylogenetic analysis of the novel Pseudomonas phage PPSC2. Adv Virol 163:1977–1980
Xuan G, Lu C, Xu H, Chen Z, Li K, Liu H, Liu H, Xia Y, Xun L (2020) Sulfane Sulfur is an intrinsic signal activating MexR-regulated antibiotic resistance in Pseudomonas aeruginosa. Mol Microbiol 114:1038–1048
Xuan G, Kong J, Wang Y, Lin H, Wang J (2023) Characterization of the newly isolated Pseudomonas phage vB_Pae_LC3I3. Virus Res 323:198978
Yu X, Xu J, Gu Y, Zhang R, Zhu Y, Liu X (2020) Molecular characterization and comparative genomic analysis of vB_PaeP_YA3, a novel temperate bacteriophage of Pseudomonas aeruginosa. Front Microbiol 11:947
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644
Zhang H, Fouts DE, DePew J, Stevens RH (2013) Genetic modifications to temperate Enterococcus faecalis phage phi Ef1 1 that abolish the establishment of lysogeny and sensitivity to repressor, and increase host range and productivity of lytic infection. Microbiology-Sgm 159:1023–1035
Acknowledgements
We thank Prof. Chao Gao (Shandong university) for strain P. aeruginosa PA14 and Mengzhe Li (Beijing University of Chemical Technology) for analyzing the terminal sequences of the genome.
Funding
This work was supported by the National Natural Science Foundation of China (No. 32201982 & No. 32270152), Natural Science Foundation of Shandong Province of China (ZR2022QC039).
Author information
Authors and Affiliations
Contributions
JK: resources, formal analysis, validation. GX: conceptualization, formal analysis, writing—original draft, HL: visualization, investigation. JW: Writing—review & editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kong, J., Xuan, G., Lin, H. et al. Characterization of a novel phage vB_Pae_HB2107-3I that infects Pseudomonas aeruginosa. Mol Genet Genomics 298, 1037–1044 (2023). https://doi.org/10.1007/s00438-023-02037-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-023-02037-x