Abstract
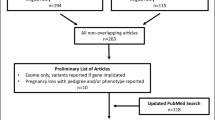
Recurrent pregnancy loss (RPL) and implantation failure (RIF) are obstacles to livebirth and multifactorial conditions in which nearly half of the cases remain unexplained, and we aimed to identify maternal candidate gene variants and pathways for RPL and RIF by analyzing whole-exome sequencing (WES) data via a new detailed bioinformatics approach. A retrospective cohort study was applied to 35 women with normal chromosomal configuration diagnosed with unexplained RPL and/or RIF. WES and comprehensive bioinformatics analyses were performed. Published gene expression datasets (n = 46) were investigated for candidate genes. Variant effects on protein structure were analyzed for 12 proteins, and BUB1B was visualized in silico. WES and bioinformatics analyses are effective and applicable for studying URPL and RIF to detect mutations, as we suggest new candidates to explain the etiology. Forty-three variants in 39 genes were detected in 29 women, 7 of them contributing to oligogenic inheritance. These genes were related to implantation, placentation, coagulation, metabolism, immune system, embryological development, cell cycle-associated processes, and ovarian functions. WES, genomic variant analyses, expression data, and protein configuration studies offer new and promising ways to investigate the etiology of URPL and RIF. Discovering etiology-identifying genetic factors can help manage couples’ needs and develop personalized therapies and new pharmaceutical products in the future. The classical approach with chromosomal analysis and targeted gene panel testing is insufficient in these cases; the exome data provide a promising way to detect and understand the possible clinical effects of the variant and its alteration on protein structure.





Similar content being viewed by others
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author. The analyzed GEO datasets underlying this article and its online supplementary material are available in the paper.
References
Alfieri C, Tischer T, Barford D (2020) A unique binding mode of Nek2A to the APC/C allows its ubiquitination during prometaphase. EMBO Rep 21:e49831. https://doi.org/10.15252/embr.201949831
Assou S, Boumela I, Haouzi D et al (2012) Transcriptome analysis during human trophectoderm specification suggests new roles of metabolic and epigenetic genes. PLoS ONE 7:e39306. https://doi.org/10.1371/journal.pone.0039306
Atasever M, Soyman Z, Demirel E et al (2016) Diminished ovarian reserve: is it a neglected cause in the assessment of recurrent miscarriage? A cohort study. Fertil Steril 105:1236–1240. https://doi.org/10.1016/j.fertnstert.2016.01.001
Avbelj Stefanija M, Jeanpierre M, Sykiotis GP et al (2012) An ancient founder mutation in PROKR2 impairs human reproduction. Hum Mol Genet 21:4314–4324. https://doi.org/10.1093/hmg/dds264
Bajnóczky K, Gardó S (1993) “Premature anaphase” in a couple with recurrent miscarriages. Hum Genet 92:388–390. https://doi.org/10.1007/BF01247341
Barrett T, Wilhite SE, Ledoux P et al (2013) NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 41:D991-995. https://doi.org/10.1093/nar/gks1193
Baumgarten SC, Convissar SM, Zamah AM et al (2015) FSH Regulates IGF-2 expression in human granulosa cells in an AKT-dependent manner. J Clin Endocrinol Metab 100:E1046-1055. https://doi.org/10.1210/jc.2015-1504
Bell CE, Calder MD, Watson AJ (2008) Genomic RNA profiling and the programme controlling preimplantation mammalian development. Mol Hum Reprod 14:691–701. https://doi.org/10.1093/molehr/gan063
Bender Atik R, Christiansen OB, Elson J et al (2018) ESHRE guideline: recurrent pregnancy loss. Hum Reprod Open. https://doi.org/10.1093/hropen/hoy004
Bolanos-Garcia VM, Lischetti T, Matak-Vinković D et al (2011) Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding Site. Structure 19:1691–1700. https://doi.org/10.1016/j.str.2011.09.017
Bondeson M-L, Ericson K, Gudmundsson S et al (2017) A nonsense mutation in CEP55 defines a new locus for a Meckel-like syndrome, an autosomal recessive lethal fetal ciliopathy. Clin Genet 92:510–516. https://doi.org/10.1111/cge.13012
Bøtkjær JA, Pors SE, Petersen TS et al (2019) Transcription profile of the insulin-like growth factor signaling pathway during human ovarian follicular development. J Assist Reprod Genet 36:889–903. https://doi.org/10.1007/s10815-019-01432-x
Cannon J, Van Steijvoort E, Borry P, Chokoshvili D (2019) How does carrier status for recessive disorders influence reproductive decisions? A systematic review of the literature. Expert Rev Mol Diagn 19:1117–1129. https://doi.org/10.1080/14737159.2020.1690456
Capalbo A, Valero RA, Jimenez-Almazan J et al (2019) Optimizing clinical exome design and parallel gene-testing for recessive genetic conditions in preconception carrier screening: Translational research genomic data from 14,125 exomes. PLoS Genet 15:e1008409. https://doi.org/10.1371/journal.pgen.1008409
Caronia LM, Martin C, Welt CK et al (2011) A genetic basis for functional hypothalamic amenorrhea. N Engl J Med 364:215–225. https://doi.org/10.1056/NEJMoa0911064
Copp AJ, Greene NDE, Murdoch JN (2003) The genetic basis of mammalian neurulation. Nat Rev Genet 4:784–793. https://doi.org/10.1038/nrg1181
Coughlan C, Ledger W, Wang Q et al (2014) Recurrent implantation failure: definition and management. Reprod Biomed Online 28:14–38. https://doi.org/10.1016/j.rbmo.2013.08.011
Coulam CB, Roussev RG (2003) Correlation of NK cell activation and inhibition markers with NK cytoxicity among women experiencing immunologic implantation failure after in vitro fertilization and embryo transfer. J Assist Reprod Genet 20:58–62. https://doi.org/10.1023/a:1021736007376
Cristofoli F, De Keersmaecker B, De Catte L et al (2017) Novel STIL compound heterozygous mutations cause severe fetal microcephaly and centriolar lengthening. Mol Syndromol 8:282–293. https://doi.org/10.1159/000479666
D’Arcy S, Davies OR, Blundell TL, Bolanos-Garcia VM (2010) Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J Biol Chem 285:14764–14776. https://doi.org/10.1074/jbc.M109.082016
Dafinger C, Liebau MC, Elsayed SM et al (2011) Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest 121:2662–2667. https://doi.org/10.1172/JCI43639
Davis EE, Zhang Q, Liu Q et al (2011) TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43:189–196. https://doi.org/10.1038/ng.756
Delatycki MB, Alkuraya F, Archibald A et al (2020) International perspectives on the implementation of reproductive carrier screening. Prenat Diagn 40:301–310. https://doi.org/10.1002/pd.5611
Demir R, Kayisli UA, Seval Y et al (2004) Sequential expression of VEGF and its receptors in human placental villi during very early pregnancy: differences between placental vasculogenesis and angiogenesis. Placenta 25:560–572. https://doi.org/10.1016/j.placenta.2003.11.011
DePamphilis ML, Kaneko KJ, Vassilev A (2002) Activation of zygotic gene expression in mammals. In: Advances in developmental biology and biochemistry. Elsevier, pp 55–84. https://www.sciencedirect.com/science/article/pii/S1569179902120247
Dhaenens L, Lierman S, De Clerck L et al (2019) Endometrial stromal cell proteome mapping in repeated implantation failure and recurrent pregnancy loss cases and fertile women. Reprod Biomed Online 38:442–454. https://doi.org/10.1016/j.rbmo.2018.11.022
Dodé C, Teixeira L, Levilliers J et al (2006) Kallmann syndrome: mutations in the genes encoding prokineticin-2 and prokineticin receptor-2. PLoS Genet 2:e175. https://doi.org/10.1371/journal.pgen.0020175
Dohrn N, Le VQ, Petersen A et al (2015) ECEL1 mutation causes fetal arthrogryposis multiplex congenita. Am J Med Genet A 167A:731–743. https://doi.org/10.1002/ajmg.a.37018
Dória S, Sousa M, Fernandes S et al (2010) Gene expression pattern of IGF2, PHLDA2, PEG10 and CDKN1C imprinted genes in spontaneous miscarriages or fetal deaths. Epigenetics 5:444–450. https://doi.org/10.4161/epi.5.5.12118
Ellard S, Kivuva E, Turnpenny P et al (2015) An exome sequencing strategy to diagnose lethal autosomal recessive disorders. Eur J Hum Genet 23:401–404. https://doi.org/10.1038/ejhg.2014.120
Falco G, Lee S-L, Stanghellini I et al (2007) Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 307:539–550. https://doi.org/10.1016/j.ydbio.2007.05.003
Filges I, Nosova E, Bruder E et al (2014) Exome sequencing identifies mutations in KIF14 as a novel cause of an autosomal recessive lethal fetal ciliopathy phenotype. Clin Genet 86:220–228. https://doi.org/10.1111/cge.12301
Garrido-Gómez T, Quiñonero A, Antúnez O et al (2014) Deciphering the proteomic signature of human endometrial receptivity. Hum Reprod 29:1957–1967. https://doi.org/10.1093/humrep/deu171
Green RC, Berg JS, Grody WW et al (2013) ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15:565–574. https://doi.org/10.1038/gim.2013.73
Grewal T, Hoque M, Conway JRW et al (2017) Annexin A6-A multifunctional scaffold in cell motility. Cell Adh Migr 11:288–304. https://doi.org/10.1080/19336918.2016.1268318
Guzmán MA, Navarro MA, Carnicer R et al (2006) Cystathionine beta-synthase is essential for female reproductive function. Hum Mol Genet 15:3168–3176. https://doi.org/10.1093/hmg/ddl393
Hoffmann P, Saoudi Y, Benharouga M et al (2009) Role of EG-VEGF in human placentation: physiological and pathological implications. J Cell Mol Med 13:2224–2235. https://doi.org/10.1111/j.1582-4934.2008.00554.x
Huang W, Yandell BS, Khatib H (2010) Transcriptomic profiling of bovine IVF embryos revealed candidate genes and pathways involved in early embryonic development. BMC Genom 11:23. https://doi.org/10.1186/1471-2164-11-23
Inagaki A, Nishizawa H, Ota S et al (2012) Upregulation of HtrA4 in the placentas of patients with severe preeclampsia. Placenta 33:919–926. https://doi.org/10.1016/j.placenta.2012.08.003
Ji S, You Y, Kerner J et al (2008) Homozygous carnitine palmitoyltransferase 1b (muscle isoform) deficiency is lethal in the mouse. Mol Genet Metab 93:314–322. https://doi.org/10.1016/j.ymgme.2007.10.006
Jiao Z-X, Woodruff TK (2013) Detection and quantification of maternal-effect gene transcripts in mouse second polar bodies: potential markers of embryo developmental competence. Fertil Steril 99:2055–2061. https://doi.org/10.1016/j.fertnstert.2013.02.003
Kaback M, Lim-Steele J, Dabholkar D et al (1993) Tay-Sachs disease–carrier screening, prenatal diagnosis, and the molecular era. An international perspective, 1970 to 1993. The International TSD Data Collection Network. JAMA 270:2307–2315
Kirshenbaum M, Orvieto R (2019) Should we offer in vitro fertilization to couples with unexplained recurrent pregnancy loss? J Clin Med 8:E2001. https://doi.org/10.3390/jcm8112001
Larsen EC, Christiansen OB, Kolte AM, Macklon N (2013) New insights into mechanisms behind miscarriage. BMC Med 11:154. https://doi.org/10.1186/1741-7015-11-154
Lavery GG, Walker EA, Tiganescu A et al (2008) Steroid biomarkers and genetic studies reveal inactivating mutations in hexose-6-phosphate dehydrogenase in patients with cortisone reductase deficiency. J Clin Endocrinol Metab 93:3827–3832. https://doi.org/10.1210/jc.2008-0743
Li L, Zheng P, Dean J (2010) Maternal control of early mouse development. Development 137:859–870. https://doi.org/10.1242/dev.039487
Lucotte G, Mercier G (2001) Population genetics of factor V Leiden in Europe. Blood Cells Mol Dis 27:362–367. https://doi.org/10.1006/bcmd.2001.0388
Maddirevula S, Awartani K, Coskun S et al (2020) A genomics approach to females with infertility and recurrent pregnancy loss. Hum Genet 139:605–613. https://doi.org/10.1007/s00439-020-02143-5
Mascarenhas MN, Flaxman SR, Boerma T et al (2012) National, regional, and global trends in infertility prevalence since 1990: a systematic analysis of 277 health surveys. PLoS Med 9:e1001356. https://doi.org/10.1371/journal.pmed.1001356
Nuño-Ayala M, Guillén N, Arnal C et al (2012) Cystathionine β-synthase deficiency causes infertility by impairing decidualization and gene expression networks in uterus implantation sites. Physiol Genom 44:702–716. https://doi.org/10.1152/physiolgenomics.00189.2010
Patrizio P, Fragouli E, Bianchi V et al (2007) Molecular methods for selection of the ideal oocyte. Reprod Biomed Online 15:346–353. https://doi.org/10.1016/s1472-6483(10)60349-5
Perrier d’Hauterive S, Charlet-Renard C, Berndt S et al (2004) Human chorionic gonadotropin and growth factors at the embryonic-endometrial interface control leukemia inhibitory factor (LIF) and interleukin 6 (IL-6) secretion by human endometrial epithelium. Hum Reprod 19:2633–2643. https://doi.org/10.1093/humrep/deh450
Pils S, Promberger R, Springer S et al (2016) Decreased ovarian reserve predicts inexplicability of recurrent miscarriage? A retrospective analysis. PLoS ONE 11:e0161606. https://doi.org/10.1371/journal.pone.0161606
Putoux A, Thomas S, Coene KLM et al (2011) KIF7 mutations cause fetal hydrolethalus and acrocallosal syndromes. Nat Genet 43:601–606. https://doi.org/10.1038/ng.826
Qiao Y, Wen J, Tang F et al (2016) Whole exome sequencing in recurrent early pregnancy loss. Mol Hum Reprod 22:364–372. https://doi.org/10.1093/molehr/gaw008
Quintero-Ronderos P, Mercier E, Fukuda M et al (2017) Novel genes and mutations in patients affected by recurrent pregnancy loss. PLoS ONE 12:e0186149. https://doi.org/10.1371/journal.pone.0186149
Reynaud R, Jayakody SA, Monnier C et al (2012) PROKR2 variants in multiple hypopituitarism with pituitary stalk interruption. J Clin Endocrinol Metab 97:E1068-1073. https://doi.org/10.1210/jc.2011-3056
Salker M, Teklenburg G, Molokhia M et al (2010) Natural selection of human embryos: impaired decidualization of endometrium disables embryo-maternal interactions and causes recurrent pregnancy loss. PLoS ONE 5:e10287. https://doi.org/10.1371/journal.pone.0010287
Schäffer AA (2013) Digenic inheritance in medical genetics. J Med Genet 50:641–652. https://doi.org/10.1136/jmedgenet-2013-101713
Semczuk M, Borczynska A, Bialas M et al (2013) Expression of genes coding for proangiogenic factors and their receptors in human placenta complicated by pre-eclampsia and intrauterine growth restriction. Reprod Biol 13:133–138. https://doi.org/10.1016/j.repbio.2013.03.004
Stals KL, Wakeling M, Baptista J et al (2018) Diagnosis of lethal or prenatal-onset autosomal recessive disorders by parental exome sequencing. Prenat Diagn 38:33–43. https://doi.org/10.1002/pd.5175
Su M-T, Lin S-H, Chen Y-C, Kuo P-L (2014) Gene-gene interactions and gene polymorphisms of VEGFA and EG-VEGF gene systems in recurrent pregnancy loss. J Assist Reprod Genet 31:699–705. https://doi.org/10.1007/s10815-014-0223-2
Sutovsky P, Schatten G (2000) Paternal contributions to the mammalian zygote: fertilization after sperm-egg fusion. Int Rev Cytol 195:1–65. https://doi.org/10.1016/s0074-7696(08)62703-5
Tissir F, De-Backer O, Goffinet AM, Lambert de Rouvroit C (2002) Developmental expression profiles of Celsr (Flamingo) genes in the mouse. Mech Dev 112:157–160. https://doi.org/10.1016/s0925-4773(01)00623-2
Traboulsi W, Brouillet S, Sergent F et al (2015) Prokineticins in central and peripheral control of human reproduction. Horm Mol Biol Clin Investig 24:73–81. https://doi.org/10.1515/hmbci-2015-0040
Vettraino IM, Roby J, Tolley T, Parks WC (1996) Collagenase-I, stromelysin-I, and matrilysin are expressed within the placenta during multiple stages of human pregnancy. Placenta 17:557–563. https://doi.org/10.1016/s0143-4004(96)80072-5
Vlismas A, Bletsa R, Mavrogianni D et al (2016) Microarray analyses reveal marked differences in growth factor and receptor expression between 8-cell human embryos and pluripotent stem cells. Stem Cells Dev 25:160–177. https://doi.org/10.1089/scd.2015.0284
Vuorela P, Carpén O, Tulppala M, Halmesmäki E (2000) VEGF, its receptors and the tie receptors in recurrent miscarriage. Mol Hum Reprod 6:276–282. https://doi.org/10.1093/molehr/6.3.276
Wang T-H, Chang C-L, Wu H-M et al (2006) Insulin-like growth factor-II (IGF-II), IGF-binding protein-3 (IGFBP-3), and IGFBP-4 in follicular fluid are associated with oocyte maturation and embryo development. Fertil Steril 86:1392–1401. https://doi.org/10.1016/j.fertnstert.2006.03.064
Wang Q, Lu J, Zhang S et al (2013) Wnt6 is essential for stromal cell proliferation during decidualization in mice. Biol Reprod 88:5. https://doi.org/10.1095/biolreprod.112.104687
Watanabe M, Osada J, Aratani Y et al (1995) Mice deficient in cystathionine beta-synthase: animal models for mild and severe homocyst(e)inemia. Proc Natl Acad Sci USA 92:1585–1589. https://doi.org/10.1073/pnas.92.5.1585
Wilbe M, Ekvall S, Eurenius K et al (2015) MuSK: a new target for lethal fetal akinesia deformation sequence (FADS). J Med Genet 52:195–202. https://doi.org/10.1136/jmedgenet-2014-102730
Wilson CL, Matrisian LM (1996) Matrilysin: an epithelial matrix metalloproteinase with potentially novel functions. Int J Biochem Cell Biol 28:123–136. https://doi.org/10.1016/1357-2725(95)00121-2
Wu X, Viveiros MM, Eppig JJ et al (2003) Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet 33:187–191. https://doi.org/10.1038/ng1079
Zhang X, Nothnick WB (2005) The role and regulation of the uterine matrix metalloproteinase system in menstruating and non-menstruating species. Front Biosci 10:353–366. https://doi.org/10.2741/1533
Zhang Y, Li G, Fan Y et al (2015) Novel missense mutation in WNT6 in 100 couples with unexplained recurrent miscarriage. Hum Reprod 30:994–999. https://doi.org/10.1093/humrep/dev028
Zygmunt M, Herr F, Münstedt K et al (2003) Angiogenesis and vasculogenesis in pregnancy. Eur J Obstet Gynecol Reprod Biol 110(Suppl 1):S10-18. https://doi.org/10.1016/s0301-2115(03)00168-4
Funding
This work was supported by the Scientific Research Projects Coordination Unit of Istanbul University (Grant numbers TDK-2018-31561 and TSA-2018-32135).
Author information
Authors and Affiliations
Contributions
EGB: literature search, genomic study design, providing clinical data and blood samples, laboratory experiments, bioinformatics analyses, drafting the manuscript, preparing tables and figures, critique, and critical discussion of the manuscript. CVŞ: bioinformatics study design, bioinformatics analyses, protein structure studies, drafting the manuscript, preparing tables and figures, critique, and critical discussion of the manuscript. TK: providing clinical data and blood samples. ZOU: supervising laboratory experiments and variant analyses, critique, and critical manuscript discussion. G.BA: supervising protein structure studies and analyses, critique, and critical discussion of the manuscript, tables, and figures. SB: genomics study design, supervising the study, critique, and critical manuscript discussion.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This study was approved by the Istanbul Medical Faculty Clinical Research Ethics Committee of Istanbul University (2018-44 and 2018-249). All participants signed the informed consent form.
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Berkay, E.G., Şoroğlu, C.V., Kalaycı, T. et al. A new enrichment approach for candidate gene detection in unexplained recurrent pregnancy loss and implantation failure. Mol Genet Genomics 298, 253–272 (2023). https://doi.org/10.1007/s00438-022-01972-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01972-5