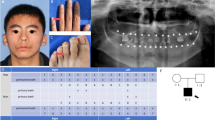
Abstract
Congenital tooth agenesis (CTA) is one of the most common craniofacial anomalies. Its frequency varies among different population depending upon the genetic heterogeneity. CTA could be of familial or sporadic and syndromic or non-syndromic. Five major genes are found to be associated with non-syndromic CTA, namely PAX9, MSX1, EDA1, AXIN2, and WNT10A. Very few studies have been carried out so far on CTA on this Indian population making this study unique and important. This study was initiated to identify potential pathogenic variant associated with congenital tooth agenesis in an India family with molar tooth agenesis. CTA was investigated and a novel c.336C > G variation was identified in the exon 3 of PAX9, leading to substitution of evolutionary conserved Cys with Trp at 112th amino acid position located at the functionally significant DNA-binding paired domain region. Functional analysis revealed that p.Cys112Trp mutation did not prevent the nuclear localization although mutant protein had higher cytoplasmic retention. EMSA using e5 probe revealed that mutant protein was unable to bind with the paired-domain-binding site. Subsequently, GST pull-down assay revealed lower binding activity of the mutant protein with its known interactor MSX1. These in vitro results were consistent with the computational results. The in vitro and computational observations altogether suggest that c.336C > G (p.Cys112Trp) variation leads to loss of function of PAX9 leading to CTA in this family.
Graphical abstract












Similar content being viewed by others
Data availability
All data included in this manuscript are available upon request by contacting with corresponding author.
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249. https://doi.org/10.1038/nmeth0410-248
Alfawaz S, Fong F, Plagnol V, Wong FS, Fearne J, Kelsell DP (2013) Recessive oligodontia linked to a homozygous loss-of-function mutation in the SMOC2 gene. Arch Oral Biol 58:462–466. https://doi.org/10.1016/j.archoralbio.2012.12.008
Arnott S, Campbell-Smith PJ, Chandrasekaran R (1976) In: Handbook of Biochemistry and Molecular Biology. CRC Press, vol 3(2), pp 411-422
Arte S, Parmanen S, Pirinen S, Alaluusua S, Nieminen P (2013) Candidate gene analysis of tooth agenesis identifies novel mutations in six genes and suggests significant role for WNT and EDA signaling and allele combinations. PLoS ONE 8:e73705. https://doi.org/10.1371/journal.pone.0073705
Bergendal B, Klar J, Stecksen-Blicks C, Norderyd J, Dahl N (2011) Isolated oligodontia associated with mutations in EDARADD, AXIN2, MSX1, and PAX9 genes. Am J Med Genet A 155a:1616–1622. https://doi.org/10.1002/ajmg.a.34045
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Bonczek O, Balcar VJ, Sery O (2017) PAX9 gene mutations and tooth agenesis: A review. Clin Genetics 92:467–476. https://doi.org/10.1111/cge.12986
Cai J, Lan Y, Appel LF, Weir M (1994) Dissection of the Drosophila paired protein: functional requirements for conserved motifs. Mech Dev 47:139–150
Das P, Hai M, Elcock C, Leal SM, Brown DT, Brook AH et al (2003) Novel missense mutations and a 288-bp exonic insertion in PAX9 in families with autosomal dominant hypodontia. Am J Med Genet A 118a:35–42. https://doi.org/10.1002/ajmg.a.10011
De Coster PJ, Marks LA, Martens LC, Huysseune A (2009) Dental agenesis: genetic and clinical perspectives. J Oral Pathol Med 38:1–17. https://doi.org/10.1111/j.1600-0714.2008.00699.x
Dugan SL, Temme RT, Olson RA, Mikhailov A, Law R, Mahmood H et al (2015) New recessive truncating mutation in LTBP3 in a family with oligodontia, short stature, and mitral valve prolapse. Am J Med Genet A 167:1396–1399. https://doi.org/10.1002/ajmg.a.37049
Garvie CW, Hagman J, Wolberger C (2001) Structural studies of Ets-1/Pax5 complex formation on DNA. Mol Cell 8:1267–1276
Hess et al (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Kantaputra P, Sripathomsawat W (2011) WNT10A and isolated hypodontia. Am J Med Genet A 155a:1119–1122. https://doi.org/10.1002/ajmg.a.33840
Kantaputra PN, Kaewgahya M, Hatsadaloi A, Vogel P, Kawasaki K, Ohazama A et al (2015) GREMLIN 2 mutations and dental anomalies. J Dent Res 94:1646–1652. https://doi.org/10.1177/0022034515608168
Kapadia H, Frazier-Bowers S, Ogawa T, D’Souza RN (2006) Molecular characterization of a novel PAX9 missense mutation causing posterior tooth agenesis. Eur J Hum Genet 14:403–409. https://doi.org/10.1038/sj.ejhg.5201574
Kiefer F, Arnold K, Künzl M, BordoliL,Schwede T (2009) The SWISS-MODEL Repository and associated resources. Nucleic acids research, 37(Database issue):D387-392. https://doi.org/10.1093/nar/gkn750
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Aguilera MA, Meyer R et al (2019) VarSome: the human genomic variant search engine. Bioinformatics 35(11):1978–1980. https://doi.org/10.1093/bioinformatics/bty897
Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I et al (2004) Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74:1043–1050. https://doi.org/10.1086/386293
Laskowski RA, Jabłońska J, Pravda L, Vařeková RS, Thornton JM (2018) PDBsum: structural summaries of PDB entries. Protein Science : a Publication of the Protein Society 27(1):129–134. https://doi.org/10.1002/pro.3289
Massink MP, Creton MA, Spanevello F, Fennis WM, Cune MS, Savelberg SM et al (2015) Loss-of-function mutations in the WNT Co-receptor LRP6 cause autosomal-dominant oligodontia. Am J Hum Genet 97:621–626. https://doi.org/10.1016/j.ajhg.2015.08.014
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405. https://doi.org/10.1093/bioinformatics/16.4.404
Mensah JK, Ogawa T, Kapadia H, Cavender AC, D’Souza RN (2004) Functional analysis of a mutation in PAX9 associated with familial tooth agenesis in humans. J Biol Chem 279:5924–5933. https://doi.org/10.1074/jbc.M305648200
Molin A, Lopez-Cazaux S, Pichon O, Vincent M, Isidor B, Le Caignec C (2015) Patients with isolated oligo/hypodontia caused by RUNX2 duplication. Am J Med Genet A 167:1386–1390. https://doi.org/10.1002/ajmg.a.37052
Nieminen P, Arte S, Tanner D, Paulin L, Alaluusua S, Thesleff I et al (2001) Identification of a nonsense mutation in the PAX9 gene in molar oligodontia. Eur J Hum Genet 9:743–746. https://doi.org/10.1038/sj.ejhg.5200715
Ogawa T, Kapadia H, Wang B, D’Souza RN (2005) Studies on Pax9-Msx1 protein interactions. Arch Oral Biol 50:141–145. https://doi.org/10.1016/j.archoralbio.2004.09.011
Ogawa T, Kapadia H, Feng JQ, Raghow R, Peters H, D’Souza RN (2006) Functional consequences of interactions between Pax9 and Msx1 genes in normal and abnormal tooth development. J Biol Chem 281:18363–18369. https://doi.org/10.1074/jbc.M601543200
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J comput chem 25(13):1605–1612
Polder BJ, Van’t Hof, M.A., Van der Linden, F.P., Kuijpers-Jagtman, A.M. (2004) A meta-analysis of the prevalence of dental agenesis of permanent teeth. Community Dent Oral Epidemiol 32:217–226. https://doi.org/10.1111/j.1600-0528.2004.00158.x
Ranjan PN, Devi C, Das P (2021) Bioinformatics analysis of SARS-CoV-2 RBD mutant variants and insights into antibody and ACE2 receptor binding. BioRxiv. https://doi.org/10.1101/2021.04.03.438113
Ranjan, P, Mohapatra, B, Das, P (2020) A rational drug designing: What bioinformatics approach tells about the wisdom of practicing traditional medicines for screening the potential of Ayurvedic and natural compounds for their inhibitory effect against COVID-19 Spike, Indian strain Spike, Papain-like protease and Main Protease protein, PREPRINT (Version 1) available at Research Square. https://doi.org/10.21203/rs.3.rs-30366/v1
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47:D886–D894. https://doi.org/10.1093/nar/gky1016.PMID:30371827;PMCID:PMC6323892
Sarkar T, Bansal R, Das P (2014) Whole genome sequencing reveals novel non-synonymous mutation in ectodysplasin A (EDA) associated with non-syndromic X-linked dominant congenital tooth agenesis. PLoS ONE 9:e106811. https://doi.org/10.1371/journal.pone.0106811
Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33(suppl_2):W363–W367
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362. https://doi.org/10.1038/nmeth.2890
Stockton DW, Das P, Goldenberg M, D’Souza RN, Patel PI (2000) Mutation of PAX9 is associated with oligodontia. Nat Genet 24:18–19. https://doi.org/10.1038/71634
Studio D (2008) Discovery studio. Accelrys [2.1]
Tao R, Jin B, Guo SZ, Qing W, Feng GY, Brooks DG et al (2006) A novel missense mutation of the EDA gene in a Mongolian family with congenital hypodontia. J Hum Genet 51:498–502. https://doi.org/10.1007/s10038-006-0389-2
Turner PJ (2005) XMGRACE, Version 5.1.19. Center for coastal and land-margin research. Oregon Graduate Institute of Science and Technology, Beaverton, OR
van den Boogaard MJ, Creton M, Bronkhorst Y, van der Hout A, Hennekam E, Lindhout D et al (2012) Mutations in WNT10A are present in more than half of isolated hypodontia cases. J Med Genet 49:327–331. https://doi.org/10.1136/jmedgenet-2012-100750
Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE (1996) A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet 13:417–421. https://doi.org/10.1038/ng0896-417
Wang Y, Groppe JC, Wu J, Ogawa T, Mues G, D’Souza RN et al (2009a) Pathogenic mechanisms of tooth agenesis linked to paired domain mutations in human PAX9. Hum Mol Genet 18:2863–2874. https://doi.org/10.1093/hmg/ddp221
Wang Y, Wu H, Wu J, Zhao H, Zhang X, Mues G et al (2009b) Identification and functional analysis of two novel PAX9 mutations. Cells Tissues Organs 189:80–87. https://doi.org/10.1159/000151448
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534. https://doi.org/10.1016/j.bpj.2011.10.024
Xu W, Rould MA, Jun S, Desplan C, Pabo CO (1995) Crystal structure of a paired domain-DNA complex at 2.5 a resolution reveals structural basis for Pax developmental mutations. Cell 80:639–650
Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO (1999) Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev 13:1263–1275
Xue LC, Rodrigues JP, Kastritis PL, Bonvin AM, Vangone A (2016) PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32(23):3676–3678. https://doi.org/10.1093/bioinformatics/btw514 (Epub 2016 Aug 8 PMID: 27503228)
Yu P, Yang W, Han D, Wang X, Guo S, Li J et al (2016) Mutations in WNT10B Are Identified in Individuals with Oligodontia. Am J Hum Genet 99:195–201. https://doi.org/10.1016/j.ajhg.2016.05.012
Acknowledgements
We are thankful to all the members of the DEN14 family and all control individuals who provided their valuable time to participate in this study. Prof. S. Ganesh, Department of Biological Science and Bioengineering, IIT Kanpur, India, for his kind gift of pCDNA3.1 ( +) Myc-His(A) vector. We are grateful to Department of Biotechnology, Govt. of India for providing funds for this study. TS acknowledges the receipt of Junior and Senior Research Fellowship from University Grants Commission, Govt. of India (http://www.ugc.ac.in/).
Funding
This study was supported by The Department of Biotechnology, Govt. of India (Grant Number: BT/PR12638/MED/12/467/2009 dated 18/03/2010; http://dbtindia.nic.in/index.asp).
Author information
Authors and Affiliations
Contributions
P.D. and T.S. conceived and planned the experiments. S.K. identified and performed clinical investigations. A.G. planned the experiment for GST pull-down. P.D. supervised and arranged funding. P.R. did bioinformatics analysis. T.S. performed all the experiments. P.D. and T.S. wrote the manuscript. T.S., S.K., A.G., and P.D. critically reviewed the findings and the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have any financial relationship and declare no conflict of interest.
Additional information
Communicated by Shuhua Xu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sarkar, T., Ranjan, P., Kanathur, S. et al. An in vitro and computational validation of a novel loss-of-functional mutation in PAX9 associated with non-syndromic tooth agenesis. Mol Genet Genomics 298, 183–199 (2023). https://doi.org/10.1007/s00438-022-01970-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01970-7