Abstract
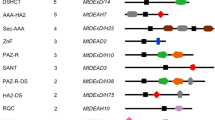
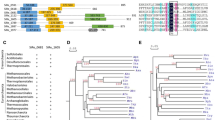
Owing to their crucial role in genome maintenance, RecQ helicases are ubiquitous and present across organisms. Though the multiplicity of RecQ helicases is well known in higher organisms, it is rare among bacteria. The ancient cyanobacterium Nostoc sp. strain PCC7120 was found to have three annotated RecQ helicases. This study aims at understanding its structural differences and evolution through bioinformatics approach and functionality through expression analysis studies. Nostoc RecQ helicases were found to be transcriptionally regulated by LexA and DNA damage inducing stresses. Bioinformatic analysis revealed that all three RecQ helicases of Nostoc possess helicases_C and Zn+2-binding domains. Two of the helicases (AnRecQ and AnRecQ2) lacked the complete RQC and HRDC domains, and AnRecQ2 had an additional Phosphoribosyl transferase domain (Pribosyltran), also seen in RecQ-like helicase (RqlH) protein of Mycobacterium smegmatis. AnRecQ1, which was similar to most bacterial RecQ helicases, differed in having a long C-terminal tail. STRING analysis revealed that the proteins also differed in their predicted protein interactome. Phylogenetic analysis suggested that the multiple recQ genes may have been acquired through duplication and acquisition of additional domains from the smallest of the RecQ helicases (AnRecQ) to cater multiple functions required to deal with the harsh environmental conditions. In course of evolution, however, the multiplicity was lost with the modern-day bacteria and lower eukaryotes which retained fewer RecQ helicases, while further duplication of the acquired RECQ occurred in higher animals and plants to deal with cellular complexity.





Similar content being viewed by others
References
Bachrati CZ, Hickson ID (2003) RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J 374(3):577–606
Bernstein DA, Keck JL (2003) Domain mapping of Escherichia coli RecQ defines the roles of conserved N-and C-terminal regions in the RecQ family. Nucleic Acids Res 31(11):2778–2785
Bernstein DA, Zittel MC, Keck JL (2003) High-resolution structure of the E coli RecQ helicase catalytic core. EMBO J 22(19):4910–4921
Boiteux S, O’Connor TR, Lederer F, Gouyette A, Laval J (1990) Homogeneous Escherichia coli FPG protein A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J Biol Chem 265(7):3916–3922
Brock TD (1973) Lower pH limit for the existence of blue-green algae: evolutionary and ecological implications. Science 179(4072):480–483
Buljubašić M, Zahradka D, Zahradka K (2013) RecQ helicase acts before RuvABC, RecG and XerC proteins during recombination in recBCD sbcBC mutants of Escherichia coli. Res Microbiol 164(10):987–997
Cai H, Jiang H, Krumholz LR, Yang Z (2014) Bacterial community composition of size-fractioned aggregates within the phycosphere of cyanobacterial blooms in a eutrophic freshwater lake. PLoS ONE 9(8):e102879
Castenholz RW (1988) Culturing methods for cyanobacteria. Method Enzymol. 167:68–93
Demoulin CF, Lara YJ, Cornet L, François C, Baurain D, Wilmotte A, Javaux EJ (2019) Cyanobacteria evolution: insight from the fossil record. Free Rad Biol Med 140:206–223
Fernández S, Sorokin A, Alonso JC (1998) Genetic recombination in Bacillus subtilis 168: effects of recu and recs mutations on DNA repair and homologous recombination. J Bacteriol 180(13):3405–3409
Frei C, Gasser SM (2000) RecQ-like helicases: the DNA replication checkpoint connection. J Cell Sci 113(15):2641–2646
Guarné A (2012) The functions of MutL in mismatch repair: the power of multitasking. Prog Mol Biol Transl Sci 110:41–70
Guo RB, Rigolet P, Zargarian L, Fermandjian S, Xi XG (2005) Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom’s syndrome helicase. Nucleic Acids Res 33(10):3109–3124
Hartung F, Puchta H (2006) The RecQ gene family in plants. J Plant Physiol 163(3):287–296
Hishida T, Han YW, Shibata T, Kubota Y, Ishino Y, Iwasaki H, Shinagawa H (2004) Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replication forks. Genes Dev 18(15):1886–1897
Huang LF, Zhang SW, Hua XT, Gao GJ, Hua YJ (2006) Construction of the recQ double mutants and analysis of adversity in Deinococcus radiodurans. Acta Microbiol Sin 46(2):205–209
Huber MD, Duquette ML, Shiels JC, Maizels N (2006) A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol 358(4):1071–1080
Karow JK, Wu L, Hickson ID (2000a) RecQ family helicases: roles in cancer and aging. Curr Opin Genet Dev 10(1):32–38
Karow JK, Constantinou A, Li JL, West SC, Hickson ID (2000b) The Bloom’s syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA 97(12):6504–6508
Killoran MP, Keck JL (2006) Three HRDC domains differentially modulate Deinococcus radiodurans RecQ DNA helicase biochemical activity. J Biol Chem 281(18):12849–12857
Killoran MP, Keck J (2008) Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic Acids Res 36(9):3139–3149
Kirti A, Rajaram H, Apte SK (2013) Characterization of two naturally truncated, Ssb-like proteins from the nitrogen-fixing cyanobacterium, Anabaena sp. PCC7120. Photosyn Res 118(1–2):147–154
Kumar A, Kirti A, Rajaram H (2018a) Regulation of multiple abiotic stress tolerance by LexA in the cyanobacterium Anabaena sp. strain PCC7120. Biochim Biophys Acta Gene Reg Mech 1861:864–877
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018b) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549
Lee JW, Harrigan J, Opresko PL, Bohr VA (2005) Pathways and functions of the Werner syndrome protein. Mech Ageing Dev 126(1):79–86
Liu JL, Rigolet P, Dou S, Wang PY, Xi XG (2004) The zinc finger motif of Escherichia coli RecQ is implicated in both DNA binding and protein folding. J Biol Chem 279(41):42794–42802
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods 25(4):402–408
Lu H, Davis AJ (2021) Human RecQ helicases in DNA double-strand break repair. Front Cell Dev Biol 9:640755
Mackinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140(2):315–322
Morimatsu K, Kowalczykowski SC (2014) RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc Natl Acad Sci USA 111(48):E5133–E5142
Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt PC (1984) Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet 195(3):474–480
Ordonez H, Unciuleac M, Shuman S (2012) Mycobacterium smegmatis RqlH defines a novel clade of bacterial RecQ-like DNA helicases with ATP-dependent 3′-5′ translocase and duplex unwinding activities. Nucleic Acids Res 40(10):4604–4614
Pandey S, Kirti A, Kumar A, Rajaram H (2018) The SbcC and SbcD homologs of the cyanobacterium Anabaena sp. strain PCC7120 (Alr3988 and All4463) contribute independently to DNA repair. Funct Integr Genom 18(4):357–367
Pandey S, Kumar A, Kirti A, Gupta GD, Rajaram H (2021) Rec (F/O/R) proteins of the nitrogen-fixing cyanobacterium Nostoc PCC7120: in silico and expression analysis. Gene 788:145663
Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A (2008) The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J Biol Chem 283(26):17766–17776
Rajaram H, Apte SK (2010) Differential regulation of groESL operon expression in response to heat and light in Anabaena. Arch Microbiol 192(9):729–738
Rajaram H, Kumar A, Kirti A, Pandey S (2020) Double strand break (DSB) repair in cyanobacteria: understanding the process in an ancient organism. DNA Repair 95:102942
Sanchez H, Kidane D, Reed P, Curtis FA, Cozar MC, Graumann PL, Sharples GJ, Alonso JC (2005) The RuvAB branch migration translocase and RecU Holliday junction resolvase are required for double-stranded DNA break repair in Bacillus subtilis. Genetics 171(3):873–883
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
Singleton MR, Wigley DB (2002) Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol 184(7):1819–1826
Singleton MR, Dillingham MS, Wigley DB (2007) Structure and mechanism of helicases and nucleic acid translocases. Ann Rev Biochem 76:23–50
Sun H, Karow JK, Hickson ID, Maizels N (1998) The Bloom’s syndrome helicase unwinds G4 DNA. J Biol Chem 273(42):27587–27592
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613
Teng FY, Wang TT, Guo HL, Xin BG, Sun B, Dou SX, Xi XG, Hou XM (2020) The HRDC domain oppositely modulates the unwinding activity of E coli RecQ helicase on duplex DNA and G-quadruplex. J Biol Chem 295(51):17646–17658
Umezu K, Nakayama K, Nakayama H (1990) Escherichia coli RecQ protein is a DNA helicase. Proc Natl Acad Sci USA 87(14):5363–5367
Vindigni A, Hickson ID (2009) RecQ helicases: multiple structures for multiple functions? HFSP Journal 3(3):153–164. https://doi.org/10.2976/1.3079540
von Kobbe C, Thomä NH, Czyzewski BK, Pavletich NP, Bohr VA (2003) Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem 278(52):52997–53006
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics (oxford, England) 25(9):1189–1191
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer T, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303
Xu HQ, Deprez E, Zhang AH, Tauc P, Ladjimi MM, Brochon JC, Auclair C, Xi XG (2003) The Escherichia coli RecQ helicase functions as a monomer. J Biol Chem 278(37):34925–34933
Yasmin T, Azeroglu B, Cockram CA, Leach DRF (2021) Distribution of Holliday junctions and repair forks during Escherichia coli DNA double-strand break repair. PLoS Genet 17(8):e1009717
Zhang XD, Dou SX, Xie P, Hu JS, Wang PY, Xi XG (2006) Escherichia coli RecQ is a rapid, efficient and monomeric helicase. J Biol Chem 281(18):12655–12663
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, A., Rajaram, H. Insights into the presence of multiple RecQ helicases in the ancient cyanobacterium, Nostoc sp. strain PCC7120: bioinformatics and expression analysis approach. Mol Genet Genomics 298, 37–47 (2023). https://doi.org/10.1007/s00438-022-01963-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01963-6