Abstract
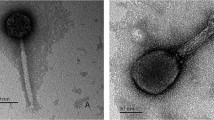
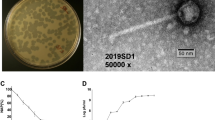
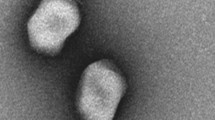
In the context of widespread bacterial contamination and the endless emergence of antibiotic-resistant bacteria, more effective ways to control pathogen infection are urgently needed. Phages become potential bactericidal agents due to their bactericidal specificity and not easy resistance to bacteria. But an important factor limiting its development is the lack of phage species. Therefore, the isolation of more new phages and studying their biological and genomic characteristics is of great significance for subsequent applications. So, in this study, SGF3, a Microviridae phage, which has shown lytic activity against Shigella flexneri, was isolated, purified, and characterized. Morphological and phylogenetic analyses identified it as a phiX174 species belonging to the Microviridae family. The latent period of phage SGF3 was 20 min, with an average burst size of approximately 7.1. Host spectrum experiments indicated its strong host specificity. Furthermore, the biofilm removal efficiency was increased by 20%-25% when SGF3 was coupled with other phages. In conclusion, the phage SGF3 found in this study was a lytic phage belonging to the Microviral family, and could be added as an auxiliary material in the phage cocktail. Studies of its characteristics and bactericidal properties had enriched the germplasm resources of microphages, provided more potential material in fighting against emerging and existing multidrug-resistant bacteria.





Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article.
Code availability
All software or custom code used during this study are available.
References
Abedon ST, Kuhl SJ, Blasdel BG, Kutter EM (2011) Phage treatment of human infections. Bacteriophage 1:66–85
APH Association (2000) Control of communicable diseases manual. J Env Health 63(5):58–60
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) Spades: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, Mckenna R et al (2002) Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol 4(184):1089–1094
Broberg CA, Palacios M, Miller VL (2013) Whole-genome draft sequences of three multidrug-resistant klebsiella pneumoniae strains available from the American type culture collection. Genome Announc 1(3):e00312-e313
Burch AD, Fane BA (2000) Foreign and chimeric external scaffolding proteins as inhibitors of microviridae morphogenesis. J Virol 10:9347–9352
Burch AD, Fane BA (2003) Genetic analyses of putative conformation switching and cross-species inhibitory domains in Microviridae external scaffolding proteins. Virology 310(1):64–71
Cisek AA, Dąbrowska I, Gregorczyk KP, Wyżewski Z (2017) Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Curr Microb 74:277–283
Cui X, Wang J, Yang C, Liang B, Ma Q, Yi S, Li H, Liu H, Li P, Zhihao Wu, Xie J, Jia L, Hao R, Wang L, Hua Y, Qiu S, Song H (2020) Prevalence and antimicrobial resistance of shigella flexneri serotype 2 variants in China. Front Microb 6:435
Dakheel KH, Rahim RA, Neela VK, Alobaidi JR, Yusoff K (2019) Genomic analyses of two novel biofilm-degrading methicillin-resistant staphylococcus aureus phages. BMC Microb 19(1):114
Finsterbusch T, Mankertz A (2009) Porcine circoviruses-small but powerful. Virus Res 143:177–183
Flemming H-C, Wingender J (2010) The biofilm matrix. Nat Rev Microb 8:623
Ghosh S, Persad E, Shiue TY, Lam C, Islam A, Mascibroda L, Sherman M, Smith T, Cheeptham N (2018) Explorative study on isolation and characterization of a microviridae G4 bacteriophage, EMCL318, against multi-drug-resistant escherichia coli 15–318. Antibiotics 7(4):92
Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30(5):1229–1235
Khalil IA, Troeger C, Blacker BF (2018) Morbidity and mortality due to shigella and enterotoxigenic Escherichia coli diarrhoea: the Global Burden of Disease Study 1990-2016 (vol 18, pg 1229, 2018). Lancet Infect Dis 18(12):1305
Matthews R (1979) Virus taxonomy: classification and nomenclature of viruses: ninth report of the international committee on taxonomy of viruses. Intervirology. 12(3–5):129–296
Owens RA, Flores R, Di Serio F et al (2012) Viroids. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy: ninth report of the international committee on taxonomy of viruses. Elsevier/Academic Press, London, UK, pp 1221–1234
Krupovic M, Forterre P (2011) Microviridae goes temperate: microvirus-related proviruses reside in the genomes of bacteroidetes. PLoS One 6(5):e19893
Labonté JM, Hallam SJ, Suttle CA (2015) Previously unknown evolutionary groups dominate the ssDNA gokushoviruses in oxic and anoxic waters of a coastal marine environment. Front Microb 6:315
Labrie SJ, Dupuis ME, Tremblay DM, Plante PL, Corbeil J, Moineau S (2014) A new microviridae phage isolated from a failed biotechnological process driven by escherichia coli. Appl Env Microb 80(22):6992–7000
Li XH, Lee JH (2017) Antibiofilm agents: a new perspective for antimicrobial strategy. J Microb 55:753–766
Li S, Lu S, Huang H, Tan L, Ni Q, Shang W et al (2019) Comparative analysis and characterization of enterobacteria phage SSL-2009a and ‘HK578likevirus’ bacteriophages. Virus Res 259:77–84
Lili Z, Huiyan D, Khashayar S, Abbas S-D, Heye W, Tao H, Lichang S, Ruicheng W, Ran W (2022) Phage JS02, a putative temperate phage, a novel biofilm-degrading agent for Staphylococcus aureus. Lett Appl Microbiol. https://doi.org/10.21203/rs.3.rs-69317/v1
Lowe TM, Chan PP (2016) tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57
Lu H, Liu H, Lu M, Wang J, Liu X, Liu R (2020a) Isolation and characterization of a novel myovirus infecting Shigella dysenteriae from the aeration tank water. Appl Biochem Biotechnol 192(3):120–131
Lu H, Yan P, Xiong W, Wang J, Liu X (2020b) Genomic characterization of a novel virulent phage infecting Shigella flexneri and isolated from sewage. Virus Res 283:197983
Lu M, Liu H, Lu H, Liu R, Liu X (2020c) Characterization and genome analysis of a novel Salmonella phage vB_SenS_SE1. Curr Microb 77(6):1308–1315
Makovcova J, Babak V, Kulich P, Masek J, Cincarova L (2017) Dynamics of mono- and dual-species biofilm formation and interactions between Staphylococcus aureus and Gram-negative bacteria. Microb Biotechnol 10(4):819–832
Martin-Laurent F, Marti R, Waglechner N, Wright GD, Topp E (2014) Draft genome sequence of the sulfonamide antibiotic-degrading Microbacterium sp strain c448. Genome Announc 2(1):e01113-13
Mathers CD, Boerma T, Ma FD (2009) Global and regional causes of death. Br Med Bull 92:7–32
McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, Rossmann MG, Olson NH, Baker TS, Incardona NL (1992) Atomic structure of single-stranded DNA bacteriophage phiX174 and its functional implications. Nature 355:137–143
Nagar V, Godambe LP, Shashidhar R (2017) Radiation sensitivity of planktonic and biofilm-associated Shigella spp and Aeromonas spp on food and food-contact surfaces. Int J Food Sci Technol 52(1):258–265
Paule A, Frezza D, Edeas M (2018) Microbiota and phage therapy: future challenges in medicine. Med Sci. https://doi.org/10.3390/medsci6040086
Peng J, Li J, Feng H, Gaofei D, Xinzhi Lu, Yuchao G, Wengong Y (2011) Antibiofilm activity of an exopolysaccharide from marine bacterium Vibrio sp QY101. PLoS One 6(4):e18514
Pepin KM, Domsic J, McKenna R (2008) Genomic evolution in a virus under specific selection for host recognition. Infect Genet Evol 8(6):825–834
Rokyta DR, Burch CL, Caudle SB, Wichman HA (2006) Horizontal gene transfer and the evolution of microvirid coliphage genomes. J Bacteriol 188(3):1134–1142
Roux S, Krupovic M, Poulet A, Debroas D, Enault F (2012) Evolution and diversity of the microviridae viral family through a collection of 81 new complete genomes assembled from virome reads. PLoS One 7(7):e40418
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York, NY, USA
Serwer P (2017) Restoring logic and data to phage-cures for infectious disease. AIMS Microb 3(4):706–712
Stothard P, Wishart DS (2005) Circular genome visualization and exploration using CGView. Bioinf 21(4):537–539
Sulakvelidze A, Alavidze Z, Morris JG (2001) Bacteriophage therapy. Antimicrob Agents Chemother 45:649–659
Sun L, Young LN, Zhang X, Boudko SP, Fokine A, Zbornik E, Roznowski AP, Molineux IJ, Rossmann MG, Fane BA (2014) Icosahedral bacteriophage phiX174 forms a tail for DNA transport during infection. Nature 505:432–435
Wu JA, Kusuma C, Mond JJ, Kokai-Kun JF (2003) Lysostaphin disrupts staphylococcus aureus and staphylococcus epidermidis biofilms on artificial surfaces. Antimicrob Agents Chemother 47(11):3407–3414
Yesigat T, Jemal M, Birhan W (2020) Prevalence and associated risk factors of salmonella, shigella, and intestinal parasites among food handlers in motta town, North West Ethiopia. Can J Infect Dis Med Microb 77:1–11
You Z, Yongjie L, Lynch KH, Dennis JJ, Wishart DS (2011) PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352
Zhan Y, Chen F (2019) The smallest ssDNA phage infecting a marine bacterium. Env Microb 21(6):1916–1928
Acknowledgements
The authors would like to acknowledge the Institute of Microbiology, Chinese Academy of Sciences for electron microscopy.
Funding
This work was supported by the National Natural Science Foundation of China [No. 51378485].
Author information
Authors and Affiliations
Contributions
LH: Performed the experiments, analyzed the data and wrote the first draft of the paper. LXC: Helped design the experiment. XWB: Assisted in experiments. LZ: Reviewed the paper. YPH: Further reviewed and edited. LRY: Commented on article revisions. All authors had read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, H., Xiong, W., Li, Z. et al. Isolation and characterization of SGF3, a novel Microviridae phage infecting Shigella flexneri. Mol Genet Genomics 297, 935–945 (2022). https://doi.org/10.1007/s00438-022-01883-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01883-5