Abstract
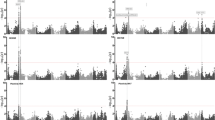
Stalk lodging severely limits the grain yield of maize (Zea mays L.). Mechanical stalk strength can be reflected by the traits of stalk diameter (SD), stalk bending strength (SBS), and lodging rind penetrometer resistance (RPR). To determine the genetic basis of maize stalk lodging, quantitative trait loci (QTLs) were mapped for these three traits using the IBM Syn10 DH population in three environments. The results indicated that there were strong genetic correlations among the three traits, and the analyses of phenotypic variations for SD, SBS, and RPR across the three environments showed high broad-sense heritability (0.6843, 0.5175, and 0.7379, respectively). In total, 44 significant QTLs were identified control the above traits across the 3 environments. A total of 14, 14, and 16 QTLs were identified for SD, SBS, and RPR across single-environment mapping, respectively. Notably, ten QTLs were stably expressed across multiple-environments, including two QTLs for SD, three for SBS, and five for RPR. Three major QTLs each accounting for over 10% of the phenotypic variation were qSD6-2 (10.03%), qSD8-2 (13.73%), and qSBS1-2 (11.89%). Comprehensive analysis of all QTLs in this study revealed that 5 QTL clusters including 12 QTLs were located on chromosomes 1, 3, 7, and 8, respectively. Among these 44 QTLs, 9 harbored 13 stalk lodging-associated SNPs that were detected by our recently published work, with 1 SNP successfully validated in the IBM Syn10 DH population. These chromosomal regions will be useful for marker-assisted selection and fine mapping of stalk lodging-related traits in maize.




Similar content being viewed by others
References
Austin D, Lee M (1998) Detection of quantitative trait loci for grain yield and yield components in maize across generations in stress and nonstress environments. Crop Sci 38(5):1296–1308
Barrière Y, Courtial A, Soler M, Grima-Pettenati J (2015) Toward the identification of genes underlying maize QTLs for lignin content, focusing on colocalizations with lignin biosynthetic genes and their regulatory MYB and NAC transcription factors. Mol Breed 35(3):87
Buckler E, Holland J, Bradbury P, Acharya C, Brown P, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz J, Goodman M, Harjes C, Guill K, Kroon D, Larsson S, Lepak N, Li H, Mitchell S, Pressoir G, Peiffer J, Rosas M, Rocheford T, Romay M, Romero S, Salvo S, Villeda H, Silva H, Sun Q, Tian F, Upadyayula N, Ware D, Yates H, Yu J, Zhang Z, Kresovich S, Mcmullen M (2009) The genetic architecture of maize flowering time. Science 325(5941):714–718
Burton A, Johnson J, Foerster J, Hirsch C, Buell C, Hanlon M, Kaeppler S, Brown K, Lynch J (2015) QTL mapping and phenotypic variation for root architectural traits in maize (Zea mays L.). Theor Appl Genet 128(1):93–106
Chang L, He K, Cui T, Xue J, Liu J (2017) QTL mapping and QTL × environment interaction analysis of Kernel ratio in maize (Zea mays). Chin J Agric Biotechol 25(4):517–525
Charcosset A, Gallais A (1996) Estimation of the contribution of quantitative trait loci (QTL) to the variance of a quantitative trait by means of genetic markers. Theor Appl Genet 93(8):1193–1201
Colbert T, Darrah L, Zuber M (1984) Effect of recurrent selection for stalk crushing strength on agronomic characteristics and soluble stalk solids in maize. Crop Sci 24(3):473–478
Daniel JR, Margaret J, Shien YL, Douglas DC (2017) Maize stalk lodging: morphological determinants of stalk strength. Crop Sci 57(2):925–934
Flintgarcia SA (2003) Quantitative trait locus analysis of stalk strength in four maize populations. Crop Sci 43(1):13–22
Gao M, Guo K, Yang Z, Li X (2003) Study on mechanical properties of cornstalk. Trans Chin Soc Agric Mach 34(4):47–49
Gou L, Huang J, Zhang B, Li T, Sun R, Zhao M (2007) Effects of population density on stalk lodging resistant mechanism and agronomic characteristics of maize. Acta Agron Sin 40(3):199–204
Hongo S, Sato K, Yokoyama R, Nishitani K (2012) Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the arabidopsis stem. Plant Cell 24(6):2624–2634
Hu H, Meng Y, Wang H, Liu H, Chen S (2012) Identifying quantitative trait loci and determining closely related stalk traits for rind penetrometer resistance in a high-oil maize population. Theor Appl Genet 124(8):1439–1447
Hu H, Liu W, Fu Z, Homann L, Technow F, Wang H, Song C, Li S, Melchinger A, Chen S (2013) QTL mapping of stalk bending strength in a recombinant inbred line maize population. Theor Appl Genet 126(9):2257–2266
Jansen C, Zhang Y, Liu H, Gonzalez-Portilla P, Lauter N, Kumar B, Trucillo-Silva L, Martin J, Lee M, Simcox K, Schussler J, Dhugga K, Lübberstedt T (2015) Genetic and agronomic assessment of cob traits in corn under low and normal nitrogen management conditions. Theor Appl Genet 128(7):1231–1242
Kamran M, Cui W, Ahmad I, Meng X, Zhang X, Su W, Chen J, Ahmad S, Fahad S, Han Q, Liu T (2018) Effect of paclobutrazol, a potential growth regulator on stalk mechanical strength, lignin accumulation and its relation with lodging resistance of maize. Plant Growth Regul 84(2):317–332
Kashiwagi T, Togawa E, Hirotsu N, Ishimaru K (2008) Improvement of lodging resistance with QTLs for stem diameter in rice (Oryza sativa L.). Theor Appl Genet 117(5):749–757
Kleczkowski L, Decker D, Wilczynska M (2011) UDP-sugar pyrophosphorylase: a new old mechanism for sugar activation. Plant Physiol 156(1):3–10
Knapp S, Stroup W, Ross W (1985) Exact confidence intervals for heritability on a progeny mean basis 1. Crop Sci 25(1):192–194
Kokubo A, Kuraishi S, Sakurai N (1989) Culm strength of barley: correlation among maximum bending stress, cell wall dimensions, and cellulose content. Plant Physiol 91(3):876–882
Kong X, Xie J, Wu X, Huang Y, Bao J (2005) Rapid prediction of acid detergent fiber, neutral detergent fiber, and acid detergent lignin of rice materials by near-infrared spectroscopy. J Agric Food Chem 53(8):2843–2848
Krakowsky M, Lee M, Coors J (2006) Quantitative trait loci for cell-wall components in recombinant inbred lines of maize (Zea mays L.) I: stalk tissue. Theor Appl Genet 112(4):717–726
Li C, Li Y, Sun B, Peng B, Liu C, Liu Z, Yang Z, Li Q, Tan W, Zhang Y, Wang D, Shi Y, Song Y, Wang T, Li Y (2013) Quantitative trait loci mapping for yield components and Kernel-related; traits in multiple connected RIL populations in maize. Euphytica 193(3):303–316
Liu Y, Wang L, Sun C, Zhang Z, Zheng Y, Qiu F (2014) Genetic analysis and major QTL detection for maize kernel size and weight in multi-environments. Theor Appl Genet 127(5):1019–1037
Liu H, Niu Y, Gonzalez-Portilla P, Zhou H, Wang L, Zuo T, Qin C, Tai S, Jansen C, Shen Y, Lin H, Lee M, Ware D, Zhang Z, Lübberstedt T, Pan G (2015) An ultra-high-density map as a community resource for discerning the genetic basis of quantitative traits in maize. BMC Genom 16(1):1078
Liu H, Zhang L, Wang J, Li C, Zeng X, Xie S, Zhang Y, Liu S, Hu S, Wang J, Lee M, Lubberstedt T, Zhao G (2017) Quantitative trait locus analysis for deep-sowing germination ability in the maize IBM Syn10 DH population. Front Plant Sci 8:813
Ma L, Guan Z, Zhang Z, Zhang X, Zhang Y, Zou C, Peng H, Pan G, Lee M, Shen Y, Lübberstedt T (2018) Identification of quantitative trait loci for leaf-related traits in an IBM Syn10 DH maize population across three environments. Plant Breed 137(2):127–138
Mizuno H, Kasuga S, Kawahigashi H (2018) Root lodging is a physical stress that changes gene expression from sucrose accumulation to degradation in sorghum. BMC Plant Biol 18(1):2
Ookawa T, Hobo T, Yano M, Murata K, Ando T, Miura H, Asano K, Ochiai Y, Ikeda M, Nishitani R, Ebitani T, Ozaki H, Angeles E, Hirasawa T, Matsuoka M (2010) New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat Commun 1(8):132
Peiffer J, Flint-Garcia S, Leon N, Mcmullen M, Kaeppler S, Buckler E (2013) The genetic architecture of maize stalk strength. PLoS One 8(6):e67066
Raihan M, Liu J, Huang J, Guo H, Pan Q, Yan J (2016) Multi-environment QTL analysis of grain morphology traits and fine mapping of a kernel-width QTL in Zheng58× SK maize population. Theor Appl Genet 129(8):1465–1477
Ramachandran S, Christensen H, Ishimaru Y, Dong C, Chao W, Cleary A, Chua N (2000) Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol 124(4):1637–1647
Remison SU, Dele A (1978) Relationship between lodging, morphological characters and yield of varieties of maize (Zea mays L.). J Agric Sci Camb 91(3):633–638
Saghai-Maroof M, Soliman K, Jorgensen R, Allard R (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81(24):8014–8018
Sekhon R, Lin H, Childs K, Hansey C, Buell C, Leon N, Kaeppler S (2011) Genome-wide atlas of transcription during maize development. Plant J 66(4):553–563
Sibale E, Darrah L, Zuber M (1992) Comparison of two rind penetrometers for measurement of stalk strength in maize. Maydica 37:111–114
Stojsin R, Ivanovic M, Kojic L, Stojsin D (1991) Inheritance of grain yield and several stalk characteristics significant in resistance to stalk lodging maize (Zea mays L.). Maydica 36:75–81
Sun Q, Liu X, Yang J, Liu W, Du Q, Wang H, Fu C, Li W (2018) microRNA528 affects lodging resistance of maize by regulating lignin biosynthesis under nitrogen-luxury conditions. Mol Plant 11(6):806–814
Tesso T, Ejeta G (2011) Stalk strength and reaction to infection by Macrophomina phaseolina of brown midrib maize (Zea mays) and sorghum (Sorghum bicolor). Field Crop Res 120(2):271–275
Timoshenko S, Gere J (1972) Mechanics of materials. Van Nostrand Reinhold Company, New York
Tuberosa R, Salvi S, Sanguineti M, Landi P, Maccaferri M, Conti S (2002) Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann Bot Lond 89(7):941–963
Wang S (2007) Windows QTL cartographer 2.5 (software)
Wang L, Li J, Yao G, Mu C, Meng Z, Liu D, Dai J (2012) Characterizations of resistance to stalk and root lodging in maize. J Maize Sci 20:69–74
Xu C, Gao Y, Tian B, Ren J, Meng Q, Wang P (2017) Effects of EDAH, a novel plant growth regulator, on mechanical strength, stalk vascular bundles and grain yield of summer maize at high densities. Field Crops Res 200:71–79
Yang C, Tang D, Qu J, Zhang L, Zhang L, Chen Z, Liu J (2016) Genetic mapping of QTL for the sizes of eight consecutive leaves below the tassel in maize (Zea mays L.). Theor Appl Genet 129(11):1–19
Zhang G, Wang X, Wang B, Tian Y, Li M, Nie Y, Peng Q, Wang Z (2013) Fine mapping a major QTL for kernel number per row under different phosphorus regimes in maize (Zea mays L.). Theor Appl Genet 126(6):1545–1553
Zhang Y, Liu P, Zhang X, Zheng Q, Chen M, Ge F, Li Z, Sun W, Guan Z, Liang T, Zheng Y, Tan X, Zou C, Peng H, Pan G, Shen Y (2018) Multi-locus genome-wide association study reveals the genetic architecture of stalk lodging resistance-related traits in maize. Front Plant Sci 9:611
Zhao X, Luo L, Cao Y, Liu Y, Li Y, Wu W, Lan Y, Jiang Y, Gao S, Zhang Z, Shen Y, Pan G, Lin H (2018) Genome-wide association analysis and QTL mapping reveal the genetic control of cadmium accumulation in maize leaf. BMC Genom 19(1):91
Acknowledgements
This work was supported by Major Project of Transgenic New Variety Cultivation (2016ZX08003003) and The National Natural Science Foundation of China (31871637).
Author information
Authors and Affiliations
Contributions
YS, GP, TR, TL, ML, and TW designed the experiments. YZ, THL, MC, ZZ, HL, GY, SZ, and CZ performed the analysis. YZ, YS, HP, and PL drafted the manuscript. All the authors critically revised and approval of the final version of this manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a potential conflict of interest.
Human or animal rights
This study does not include human or animal subjects.
Additional information
Communicated by S. Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Liang, T., Chen, M. et al. Genetic dissection of stalk lodging-related traits using an IBM Syn10 DH population in maize across three environments (Zea mays L.). Mol Genet Genomics 294, 1277–1288 (2019). https://doi.org/10.1007/s00438-019-01576-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-019-01576-6