Abstract
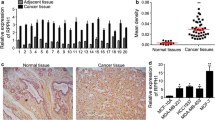
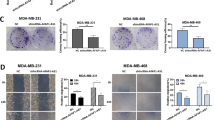
The global burden of breast cancer has been increasing, the mechanism of which is related to multifactorial accumulation of gene mutations. Dysregulation of long noncoding RNA (LncRNA) has been linked to multiple kinds of tumorigenesis. We aimed to identify functionally relevant targets of RHPN1 Antisense RNA 1 (RHPN1-AS1) in breast cancer using breast cancer cell line-based model. Quantitative RT-PCR revealed higher expression levels of RHPN1-AS1 in human breast cancer tissues and cell line MCF-7. RHPN1-AS1 was also located in MCF-7 cells by fluorescence in situ hybridization and western blot assays. Knockdown of RHPN1-AS1 delivered by lentivirus system inhibited MCF-7 cell proliferation indicated by the cell proliferation and colony formation assays, and the knockdown of RHPN1-AS1 enhanced P53 protein expression in MCF-7 and MDA-MB-231 cells. In addition, luciferase reporter assay validated that RHPN1-AS1 is a molecular sponge of miR-4261, and direct transcriptional target of c-Myc. RHPN1-AS1 exerts tumorigenesis by regulating P53 expression via MDM2 gene. These findings provide insights into the role and mechanism of action of lncRNA RHPN-AS1 in breast cancer.





Similar content being viewed by others
Abbreviations
- LncRNA:
-
Long noncoding RNA
- FISH:
-
Fluorescence in situ hybridization
References
Barr AR, Cooper S, Heldt FS, Butera F, Stoy H, Mansfeld J, Novak B, Bakal C (2017) DNA damage during S-phase mediates the proliferation-quiescence decision in the subsequent G1 via p21 expression. Nat Commun 8:14728
Benson EK, Mungamuri SK, Attie O, Kracikova M, Sachidanandam R, Manfredi JJ, Aaronson SA (2014) p53-dependent gene repression through p21 is mediated by recruitment of E2F4 repression complexes. Oncogene 33(30):3959–3969
Bodai BI, Tuso P (2015) Breast cancer survivorship: a comprehensive review of long-term medical issues and lifestyle recommendations. Perm J 19(2):48–79
Bradford JR, Cox A, Bernard P, Camp NJ (2016) Consensus analysis of whole transcriptome profiles from two breast cancer patient cohorts reveals long non-coding RNAs associated with intrinsic subtype and the tumour microenvironment. PLoS ONE 11(9):e0163238
Cerk S, Schwarzenbacher D, Adiprasito JB, Stotz M, Hutterer GC, Gerger A, Ling H, Calin GA, Pichler M (2016) Current status of long non-coding RNAs in human breast cancer. Int J Mol Sci 17(9):E1485
Coughlin SS, Ekwueme DU (2009) Breast cancer as a global health concern. Cancer Epidemiol 33(5):315–318
Danforth DN Jr (2016) Genomic changes in normal breast tissue in women at normal risk or at high risk for breast cancer. Breast Cancer 10:109–146
Dang CV (2012) MYC on the path to cancer. Cell 149(1):22–35
Ellis PA, Lonning PE, Borresen-Dale A, Aas T, Geisler S, Akslen LA, Salter I, Smith IE, Dowsett M (1997) Absence of p21 expression is associated with abnormal p53 in human breast carcinomas. Br J Cancer 76(4):480–485
Gabay M, Li Y, Felsher DW (2014) MYC activation is a hallmark of cancer initiation and maintenance. CSH Perspect Med 4(6):a014201
Goff LA, Davila J, Swerdel MR, Moore JC, Cohen RI, Wu H, Sun YE, Hart RP (2009) Ago2 immunoprecipitation identifies predicted microRNAs in human embryonic stem cells and neural precursors. PLoS ONE 4(9):e7192
Gupta GP, Massague J (2006) Cancer metastasis: building a framework. Cell 127(4):679–695
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291):1071–1076
Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387(6630):296–299
Haupt S, Vijayakumaran R, Miranda PJ, Burgess A, Lim E, Haupt Y (2017) The role of MDM2 and MDM4 in breast cancer development and prevention. J Mol Cell Biol 9(1):53–61
Huang SK, Luo Q, Peng H, Li J, Zhao M, Wang J, Gu YY, Li Y, Yuan P, Zhao GH, Huang CZ (2018) A panel of serum noncoding RNAs for the diagnosis and monitoring of response to therapy in patients with breast cancer. Med Sci Monitor 24:2476–2488
Jiao G, Huang Q, Hu M, Liang X, Li F, Lan C, Fu W, An Y, Xu B, Zhou J, Xiao J (2017) Therapeutic suppression of miR-4261 attenuates colorectal cancer by targeting MCC. Mol Ther Nucleic Acids 8:36–45
Jin X, Mu P (2015) Targeting breast cancer metastasis. Breast Cancer 9(Suppl 1):23–34
Kalkat M, De Melo J, Hickman KA, Lourenco C, Redel C, Resetca D, Tamachi A, Tu WB, Penn LZ (2017) MYC deregulation in primary human cancers. Genes 8(6):E151
Klinge CM (2018) Non-coding RNAs in breast cancer. Non-coding RNA 4(4):40
Kundu N, Brekman A, Kim JY, Xiao G, Gao C, Bargonetti J (2017) Estrogen-activated MDM2 disrupts mammary tissue architecture through a p53-independent pathway. Oncotarget 8(29):47916–47930
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Liu X, Xiao ZD, Han L, Zhang J, Lee SW, Wang W, Lee H, Zhuang L, Chen J, Lin HK, Wang J, Liang H, Gan B (2016) LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat Cell Biol 18(4):431–442
Lu L, Yu X, Zhang L, Ding X, Pan H, Wen X, Xu S, Xing Y, Fan J, Ge S, Zhang H, Jia R, Fan X (2017) The long non-coding RNA RHPN1-AS1 promotes uveal melanoma progression. Int J Mol Sci 18(1):E226
Mercer TR, Mattick JS (2013) Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol 20(3):300–307
Orom UA, Shiekhattar R (2013) Long noncoding RNAs usher in a new era in the biology of enhancers. Cell 154(6):1190–1193
Prensner JR, Chinnaiyan AM (2011) The emergence of lncRNAs in cancer biology. Cancer Discov 1(5):391–407
Sanchez-Mejias A, Tay Y (2015) Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J Hematol Oncol 8:30
Siegel RL, Miller KD, Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67(1):7–30
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30
Wong N, Wang X (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43(Database issue):D146–D152
Xu S, Kong D, Chen Q, Ping Y, Pang D (2017) Oncogenic long noncoding RNA landscape in breast cancer. Mol Cancer 16(1):129
Zhang P, Cao L, Fan P, Mei Y, Wu M (2016) LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep 17(8):1204–1220
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
All applicable international, national, and/or institutional guidelines for the care were followed.
Informed consent
Written consent has derived from each participant.
Additional information
Communicated by S. Hohmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, P., Li, Y., Li, P. et al. c-Myc induced the regulation of long non-coding RNA RHPN1-AS1 on breast cancer cell proliferation via inhibiting P53. Mol Genet Genomics 294, 1219–1229 (2019). https://doi.org/10.1007/s00438-019-01572-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-019-01572-w