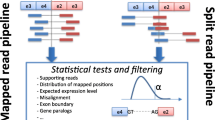
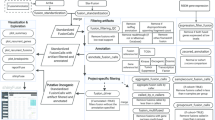
Abstract
Recurrent oncogenic fusion genes play a critical role in the development of various cancers and diseases and provide, in some cases, excellent therapeutic targets. To date, analysis tools that can identify and compare recurrent fusion genes across multiple samples have not been available to researchers. To address this deficiency, we developed Co-occurrence Fusion (Co-fuse), a new and easy to use software tool that enables biologists to merge RNA-seq information, allowing them to identify recurrent fusion genes, without the need for exhaustive data processing. Notably, Co-fuse is based on pattern mining and statistical analysis which enables the identification of hidden patterns of recurrent fusion genes. In this report, we show that Co-fuse can be used to identify 2 distinct groups within a set of 49 leukemic cell lines based on their recurrent fusion genes: a multiple myeloma (MM) samples-enriched cluster and an acute myeloid leukemia (AML) samples-enriched cluster. Our experimental results further demonstrate that Co-fuse can identify known driver fusion genes (e.g., IGH-MYC, IGH-WHSC1) in MM, when compared to AML samples, indicating the potential of Co-fuse to aid the discovery of yet unknown driver fusion genes through cohort comparisons. Additionally, using a 272 primary glioma sample RNA-seq dataset, Co-fuse was able to validate recurrent fusion genes, further demonstrating the power of this analysis tool to identify recurrent fusion genes. Taken together, Co-fuse is a powerful new analysis tool that can be readily applied to large RNA-seq datasets, and may lead to the discovery of new disease subgroups and potentially new driver genes, for which, targeted therapies could be developed. The Co-fuse R source code is publicly available at https://github.com/sakrapee/co-fuse.





Similar content being viewed by others
References
Bao ZS, Chen HM, Yang MY, Zhang CB, Yu K, Ye WL, Hu BQ, Yan W, Zhang W, Akers J, Ramakrishnan V, Li J, Carter B, Liu YW, Hu HM, Wang Z, Li MY, Yao K, Qiu XG, Kang CS, You YP, Fan XL, Song WS, Li RQ, Su XD, Chen CC, Jiang T (2014) RNA-seq of 272 gliomas revealed a novel, recurrent PTPRZ1-MET fusion transcript in secondary glioblastomas. Genome Res 24:1765–1773
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603–607
Beccuti M, Carrara M, Cordero F, Lazzarato F, Donatelli S, Nadalin F, Policriti A, Calogero RA (2014) Chimera: a bioconductor package for secondary analysis of fusion products. Bioinformatics 30:3556–3557
Bushman F (2017) Cancer Gene List. Bushman Lab. http://www.bushmanlab.org/links/genelists. Accessed 7 Feb 2017
Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW (2016) Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17:257–271
Capdeville R, Buchdunger E, Zimmermann J, Matter A (2002) Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Discov 1:493–502
Cestarelli V, Fiscon G, Felici G, Bertolazzi P, Weitschek E (2016) CAMUR: knowledge extraction from RNA-seq cancer data through equivalent classification rules. Bioinformatics 32:697–704
Cleynen A, Szalat R, Kemal Samur M, Robiou du Pont S, Buisson L, Boyle E, Chretien ML, Anderson K, Minvielle S, Moreau P, Attal M, Parmigiani G, Corre J, Munshi N, Avet-Loiseau H (2017) Expressed fusion gene landscape and its impact in multiple myeloma. Nat Commun 8:1893
Drexler HG, Dirks WG, Matsuo Y, MacLeod RA (2003) False leukemia–lymphoma cell lines: an update on over 500 cell lines. Leukemia 17:416–426
Hoogstrate Y, Bottcher R, Hiltemann S, van der Spek PJ, Jenster G, Stubbs AP (2016) FuMa: reporting overlap in RNA-seq detected fusion genes. Bioinformatics 32:1226–1228
Howe EA, Sinha R, Schlauch D, Quackenbush J (2011) RNA-Seq analysis in MeV. Bioinformatics 27:3209–3210
Jia W, Qiu K, He M, Song P, Zhou Q, Zhou F, Yu Y, Zhu D, Nickerson ML, Wan S, Liao X, Zhu X, Peng S, Li Y, Wang J, Guo G (2013) SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol 14:R12
Kim D, Salzberg SL (2011) TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 12:R72
Kuehn H, Liberzon A, Reich M, Mesirov JP (2008) Using GenePattern for gene expression analysis. Curr Protoc Bioinform. https://doi.org/10.1002/0471250953.bi0712s22
Kumar-Sinha C, Kalyana-Sundaram S, Chinnaiyan AM (2015) Landscape of gene fusions in epithelial cancers: seq and ye shall find. Genome Med 7:129
Latysheva NS, Babu MM (2016) Discovering and understanding oncogenic gene fusions through data intensive computational approaches. Nucleic Acids Res 44:4487–4503
Lee M, Lee K, Yu N, Jang I, Choi I, Kim P, Jang YE, Kim B, Kim S, Lee B, Kang J, Lee S (2017) ChimerDB 3.0: an enhanced database for fusion genes from cancer transcriptome and literature data mining. Nucleic Acids Res 45:D784-D789
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298:1912–1934
McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, Sun MG, Griffith M, Heravi Moussavi A, Senz J, Melnyk N, Pacheco M, Marra MA, Hirst M, Nielsen TO, Sahinalp SC, Huntsman D, Shah SP (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7:e1001138
Mertens F, Johansson B, Fioretos T, Mitelman F (2015) The emerging complexity of gene fusions in cancer. Nat Rev Cancer 15:371–381
Meyer C, Burmeister T, Groger D, Tsaur G, Fechina L, Renneville A, Sutton R, Venn NC, Emerenciano M, Pombo-de-Oliveira MS, Barbieri Blunck C, Almeida Lopes B, Zuna J, Trka J, Ballerini P, Lapillonne H, De Braekeleer M, Cazzaniga G, Corral Abascal L, van der Velden VHJ, Delabesse E, Park TS, Oh SH, Silva MLM, Lund-Aho T, Juvonen V, Moore AS, Heidenreich O, Vormoor J, Zerkalenkova E, Olshanskaya Y, Bueno C, Menendez P, Teigler-Schlegel A, Zur Stadt U, Lentes J, Gohring G, Kustanovich A, Aleinikova O, Schafer BW, Kubetzko S, Madsen HO, Gruhn B, Duarte X, Gameiro P, Lippert E, Bidet A, Cayuela JM, Clappier E, Alonso CN, Zwaan CM, van den Heuvel-Eibrink MM, Izraeli S, Trakhtenbrot L, Archer P, Hancock J, Moricke A, Alten J, Schrappe M, Stanulla M, Strehl S, Attarbaschi A, Dworzak M, Haas OA, Panzer-Grumayer R, Sedek L, Szczepanski T, Caye A, Suarez L, Cave H, Marschalek R (2017) The MLL recombinome of acute leukemias in 2017. Leukemia 32(2):273–284
Morgan GJ, Walker BA, Davies FE (2012) The genetic architecture of multiple myeloma. Nat Rev Cancer 12:335–348
Nicorici D, Satalan M, Edgren H, Kangaspeska S, Murumagi A, Kallioniemi O, Virtanen S, Kilkku O (2014) FusionCatcher—a tool for finding somatic fusion genes in paired-end RNA-sequencing data. BioRxiv 011650. https://doi.org/10.1101/011650
Panigrahi P, Jere A, Anamika K (2018) FusionHub: a unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS One 13:e0196588
Persson H, Sokilde R, Hakkinen J, Pirona AC, Vallon-Christersson J, Kvist A, Mertens F, Borg A, Mitelman F, Hoglund M, Rovira C (2017) Frequent miRNA-convergent fusion gene events in breast cancer. Nat Commun 8:788
Roberts KG (2017) The biology of Philadelphia chromosome-like ALL. Best Pract Res Clin Haematol 30:212–221
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, McCastlain K, Ding L, Lu C, Song G, Ma J, Becksfort J, Rusch M, Chen SC, Easton J, Cheng J, Boggs K, Santiago-Morales N, Iacobucci I, Fulton RS, Wen J, Valentine M, Cheng C, Paugh SW, Devidas M, Chen IM, Reshmi S, Smith A, Hedlund E, Gupta P, Nagahawatte P, Wu G, Chen X, Yergeau D, Vadodaria B, Mulder H, Winick NJ, Larsen EC, Carroll WL, Heerema NA, Carroll AJ, Grayson G, Tasian SK, Moore AS, Keller F, Frei-Jones M, Whitlock JA, Raetz EA, White DL, Hughes TP, Guidry Auvil JM, Smith MA, Marcucci G, Bloomfield CD, Mrozek K, Kohlschmidt J, Stock W, Kornblau SM, Konopleva M, Paietta E, Pui CH, Jeha S, Relling MV, Evans WE, Gerhard DS, Gastier-Foster JM, Mardis E, Wilson RK, Loh ML, Downing JR, Hunger SP, Willman CL, Zhang J, Mullighan CG (2014) Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371:1005–1015
Roychowdhury S, Chinnaiyan AM (2016) Translating cancer genomes and transcriptomes for precision oncology. CA Cancer J Clin 66:75–88
Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C (2014) The landscape of kinase fusions in cancer. Nat Commun 5:4846
Wang Q, Xia J, Jia P, Pao W, Zhao Z (2013) Application of next generation sequencing to human gene fusion detection: computational tools, features and perspectives. Br Bioinform 14:506–519
Weitschek E, Felici G, Bertolazzi P (2012) MALA: a microarray clustering and classification software. In: 23rd International workshop on database and expert systems applications. IEEE, Vienna, pp 201–205
Wilks C, Cline MS, Weiler E, Diehkans M, Craft B, Martin C, Murphy D, Pierce H, Black J, Nelson D, Litzinger B, Hatton T, Maltbie L, Ainsworth M, Allen P, Rosewood L, Mitchell E, Smith B, Warner J, Groboske J, Telc H, Wilson D, Sanford B, Schmidt H, Haussler D, Maltbie D (2014) The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database. https://doi.org/10.1093/database/bau093
Zhao M, Sun J, Zhao Z (2013) TSGene: a web resource for tumor suppressor genes. Nucleic Acids Res 41:D970–D976
Acknowledgements
C. H. K is a recipient of Mary Overton research fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Sakrapee Paisitkriangkrai declares that he has no conflict of interest. Kelly Quek declares that she has no conflict of interest. Eva Nievergall declares that she has no conflict of interest. Anissa Jabbour declares that she has no conflict of interest. Andrew Zannettino declares that he has no conflict of interest. Chung H Kok declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Data availability
The RNA-seq leukemic cell lines dataset analysed during the current study is publicly available at Cancer Genomics Hub (http://cghub.ucsc.edu) and NCI Genomic Data Commons (https://gdc.nci.nih.gov/). The raw sequencing data for 272 gliomas clinical samples dataset analysed during the current study is publicly available at NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE48865. The raw FusionCatcher analysis results generated during this study are included in this published article as supplementary information files.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Paisitkriangkrai, S., Quek, K., Nievergall, E. et al. Co-fuse: a new class discovery analysis tool to identify and prioritize recurrent fusion genes from RNA-sequencing data. Mol Genet Genomics 293, 1217–1229 (2018). https://doi.org/10.1007/s00438-018-1454-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-018-1454-1