Abstract
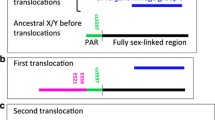
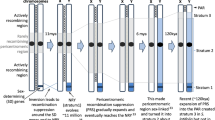
Sex chromosomes are defined by a non-recombining sex-determining region (SDR) flanked by one or two pseudoautosomal regions (PARs). The genetic composition and evolutionary dynamics of the PAR is also influenced by its linkage to the differentiated non-recombining SDR; however, understanding the effects of this linkage requires a precise definition of the PAR boundary. Here, we took a molecular population genetic approach to further refine the location of the PAR boundary of the evolutionary young sex chromosomes of the tropical plant, Carica papaya. We were able to map the position of the papaya PAR boundary A to a 100-kb region between two genetic loci approximately 2 Mb upstream of the previously genetically identified PAR boundary. Furthermore, this boundary is polymorphic within natural populations of papaya, with an approximately 100–130 kb expansion of the non-recombining SDR found in 16 % of individuals surveyed. The expansion of the PAR boundary in one Y haplotype includes at least one additional gene. Homologs of this gene are involved in male gametophyte and pollen development in other plant species.







Similar content being viewed by others
Abbreviations
- SDR:
-
Sex-determining region
- PAR:
-
Pseudoautosomal region
- QTL:
-
Quantitative trait loci
- LD:
-
Linkage disequilibrium
- MP:
-
Maximum parsimony
References
Bellott DW, Skaletsky H, Pyntikova T, Mardis ER, Graves T, Kremitzki C, Brown LG, Rozen S, Warren WC, Wilson RK et al (2010) Convergent evolution of chicken Z and human X chromosomes by expansion and gene acquisition. Nature 466:612–613
Bergero R, Charlesworth D (2009) The evolution of restricted recombination in sex chromosomes. Trends Ecol Evol 24:94–102
Bergero R, Forrest A, Kamau E, Charlesworth D (2007) Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175:1945–1954
Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D (2013) Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics 194:673–686
Blavet N, Blavet H, Cegan R, Zemp N, Zdanska J, Janousek B, Hobza R, Widmer A (2012) Comparative analysis of a plant pseudoautosomal region (PAR) in Silene latifolia with the corresponding S. vulgaris autosome. BMC Genom 13:226–235
Brown JE, Bauman JM, Lawrie JF, Rocha OJ, Moore RC (2012) The structure of morphological and genetic diversity in natural populations of Carica papaya (Caricaceae) in Costa Rica. Biotropica 44:179–188
Bussell JJ, Pearson NM, Kanda R, Filatov DA, Lahn BT (2006) Human polymorphism and human-chimpanzee divergence in pseudoautosomal region correlate with local recombination rate. Gene 368:94–100
Charlesworth D (2013) Plant sex chromosome evolution. J Exp Bot 64:405–420
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95:118–128
Charlesworth B, Jordan CY, Charlesworth D (2014) The evolutionary dynamics of sexually antagonistic mutations in pseudoautosomal regions of sex chromosomes. Evolution 68:1339–1350
Chibalina MV, Filatov DA (2011) Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol 21:1475–1479
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Delph LF, Arntz AM, Scotti-Saintagne C, Scotti I (2010) The genomic architecture of sexual dimorphism in the dioecious plant Silene latifolia. Evolution 64:2873–2886
Ellis NA, Goodfellow PJ, Pym B, Smith M, Palmer M, Frischauf AM, Goodfellow PN (1989) The pseudoautosomal boundary in man is defined by an Alu repeat sequence inserted on the Y chromosome. Nature 337:81–84
Ellis N, Yen P, Neiswanger K, Shapiro LJ, Goodfellow PN (1990) Evolution of the pseudoautosomal boundary in old-world monkeys and great apes. Cell 63:977–986
Ellis NA, Ye TZ, Patton S, German J, Goodfellow PN, Weller P (1994) Cloning of Pbdx, an Mic2-related gene that spans the pseudoautosomal boundary on chromosome Xp. Nat Genet 6:394–400
Filatov DA (2004) A gradient of silent substitution rate in the human pseudoautosomal region. Mol Biol Evol 21:410–417
Galtier N (2004) Recombination, GC-content and the human pseudoautosomal boundary paradox. Trends Gene 20:347–349
Goldberg MT, Spigler RB, Ashman TL (2010) Comparative genetic mapping points to different sex chromosomes in sibling species of wild strawberry (Fragaria). Genetics 186:1425–1433
Graves JAM, Wakefield MJ, Toder R (1998) The origin and evolution of the pseudoautosomal regions of human sex chromosomes. Hum Mol Genet 7:1991–1996
Gschwend AR, Yu QY, Tong EJ, Zeng FC, Han J, VanBuren R, Aryal R, Charlesworth D, Moore PH, Paterson AH et al (2012) Rapid divergence and expansion of the X chromosome in papaya. P Natl Acad Sci USA 109:13716–13721
Gschwend AR, Wai CM, Zee F, Arumuganathan AK, Ming R (2013) Genome size variation among sex types in dioecious and trioecious Caricaceae species. Euphytica 189:461–469
Guo SW, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportion for multiple alleles. Biometrics 48:361–372
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program from Windows 95/98/NT. Nucl Acid Symp Series 41:95–98
Hirano T, Matsuzawa T, Takegawa K, Sato MH (2011) Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol 155:797–807
Huson DH, Scornavacca C (2012) Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067
Jordan CY, Charlesworth D (2012) The potential for sexually antagonistic polymorphism in different genome regions. Evolution 66:505–516
Kipling D, Salido EC, Shapiro LJ, Cooke HJ (1996a) High frequency de novo alterations in the long-range genomic structure of the mouse pseudoautosomal region. Nat Genet 13:78–82
Kipling D, Wilson HE, Thomson EJ, Lee M, Perry J, Palmer S, Ashworth A, Cooke HJ (1996b) Structural variation of the pseudoautosomal region between and within inbred mouse strains. Proc Natl Acad Sci USA 93:171–175
Kirkpatrick M, Guerrero Scarpino (2010) Patterns of neutral genetic variation on recombining sex chromosomes. Genetics 184:1141–1152
Kirkpatrick M, Guerrero RF (2014) Signatures of sex-antagonistic selection on recombining sex chromosomes. Genetics 197:531–541
Lahn BT, Page DC (1999) Four evolutionary strata on the human X chromosome. Science 286:964–967
Lawson-Handley L, Ceplitis H, Ellegren H (2004) Evolutionary strata on the chicken Z chromosome: implications for sex chromosome evolution. Genetics 167:367–376
Leder EH, Cano JM, Leinonen T, O’Hara RB, Nikinmaa M, Primmer CR, Merila J (2010) Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol 27:1495–1503
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu ZY, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu QY, Pearl HM, Kim MS, Charlton JW, Stiles JI et al (2004) A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427:348–352
Mangs AH, Morris BJ (2007) The human pseudoautosomal region (PAR): origin, function and future. Curr Genomics 8:129–136
Manshardt RM, Zee FTP (1994) Papaya germplasm and breeding in Hawaii. Fruit Varieties J 48:146–152
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514
Ming R, Yu QY, Moore PH, Paull RE, Chen NJ, Wang ML, Zhu YJ, Schuler MA, Jiang JM, Paterson AH (2012) Genome of papaya, a fast growing tropical fruit tree. Tree Genet Genome 8:445–462
Na JK, Wang J, Murray JE, Gschwend AR, Zhang W, Yu Q, Navajas-Perez R, Feltus FA, Chen C, Kubat Z et al (2012) Construction of physical maps for the sex-specific regions of papaya sex chromosomes. BMC Genom 13:176
Nicolas M, Marais G, Hykelova V, Janousek B, Laporte V, Vyskot B, Mouchiroud D, Negrutiu I, Charlesworth D, Moneger F (2005) A gradual process of recombination restriction in the evolutionary history of the sex chromosomes in dioecious plants. PLoS Biol 3(1):e4
Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF (2011) About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet 27:358–367
Palmer S, Perry J, Kipling D, Ashworth A (1997) A gene spans the pseudoautosomal boundary in mice. P Natl Acad Sci USA 94:12030–12035
Perry J, Palmer S, Gabriel A, Ashworth A (2001) A short pseudoautosomal region in laboratory mice. Genome Res 11:1826–1832
Pigozzi MI, Solari AJ (1999) The ZW pairs of two paleognath birds from two orders show transitional stages of sex chromosome differentiation. Chromosome Res 7:541–551
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Qiu S, Bergero R, Forrest A, Kaiser VB, Charlesworth D (2010) Nucleotide diversity in Silene latifolia autosomal and sex-linked genes. P Roy Soc B Biol Sci 277:3283–3290
Qiu S, Bergero R, Charlesworth D (2013) Testing for the footprint of sexually antagonistic polymorphisms in the pseudoautosomal region of a plant sex chromosome pair. Genetics 194:663–672
Raymond M, Rousset F (1995) Genepop (version-1.2)—population-genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1984) Sex-chromosomes and the evolution of sexual dimorphism. Evolution 38:735–742
Robertson A, Hill WG (1984) Deviations from Hardy-Weinberg proportions—sampling variances and use in estimation of inbreeding coefficients. Genetics 107:703–718
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Method Mol Biol 132:365–386
Sayres MAW, Makova KD (2013) Gene survival and death on the human Y chromosome. Mol Biol Evol 30:781–787
Serrazina S, Dias FV, Malho R (2014) Characterization of FAB1 phosphatidylinositol kinases in Arabidopsis pollen tube growth and fertilization. New Phytol 203:784–793
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T et al (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837
Skinner BM, Lachani K, Sargent CA, Affara NA (2013) Regions of XY homology in the pig X chromosome and the boundary of the pseudoautosomal region. BMC Genet 14:1–7
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Telgmann-Rauber A, Jamsari A, Kinney MS, Pires JC, Jung C (2007) Genetic and physical maps around the sex-determining M-locus of the dioecious plant asparagus. Mol Genet Genomics 278:221–234
Van Laere AS, Coppieters W, Georges M (2008) Characterization of the bovine pseudoautosomal boundary: documenting the evolutionary history of mammalian sex chromosomes. Genome Res 18:1884–1895
VanBuren R, Ming R (2013) Dynamic transposable element accumulation in the nascent sex chromosomes of papaya. Mob Genet Elem 3:e23462
Vicoso B, Kaiser VB, Bachtrog D (2013) Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc Natl Acad Sci USA 110:6453–6458
Wang Y, Zhang WZ, Song LF, Zou JJ, Su Z, Wu WH (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148:1201–1211
Wang JP, Na JK, Yu QY, Gschwend AR, Han J, Zeng FC, Aryal R, VanBuren R, Murray JE, Zhang WL et al (2012) Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA 109:13710–13715
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276
Weingartner LA, Moore RC (2012) Contrasting patterns of X/Y polymorphism distinguish Carica papaya from other sex chromosome systems. Mol Biol Evol 29:3909–3920
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370
White MA, Ikeda A, Payseur BA (2012) A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm Genome 23:454–466
Whitley P, Hinz S, Doughty J (2009) Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol 151:1812–1822
Yin T, DiFazio SP, Gunter LE, Zhang X, Sewell MM, Woolbright SA, Allan GJ, Kelleher CT, Douglas CJ, Wang M et al (2008) Genome structure and emerging evidence of an incipient sex chromosome in Populus. Genome Res 18:422–430
Yu QY, Navajas-Perez R, Tong E, Robertson J, Moore PH, Paterson AH, Ming R (2008) Recent origin of dioecious and gynodioecious chromosomes in papaya. Tropical Plant Biol 1:49–57
Yu QY, Tong E, Skelton RL, Bowers JE, Jones MR, Murray JE, Hou SB, Guan PZ, Acob RA, Luo MC et al (2009) A physical map of the papaya genome with integrated genetic map and genome sequence. BMC Genom 10:371–382
Zhang WL, Wang XU, Yu QY, Ming R, Jiang JM (2008) DNA methylation and heterochromatinization in the male-specific region of the primitive Y chromosome of papaya. Genome Res 18:1938–1943
Acknowledgments
This work was supported by the National Science Foundation (DBI-0922545 to RCM) and by the Department of Botany at Miami University through Academic Challenge grants to FML. We are extremely grateful to Deborah Charlesworth and one anonymous reviewer for their critical examination of the manuscript and helpful advice. We are greatly indebted to Oscar Rocha at Kent State University for his help during the specimen collecting process in Costa Rica. We would also like to thank our collaborators Ray Ming (University of Illinois at Urbana-Champaign) and Qingyi Yu (Texas A&M University) for providing papaya sequence information and cultivar tissues.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. Gebhardt.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lappin, F.M., Medert, C.M., Hawkins, K.K. et al. A polymorphic pseudoautosomal boundary in the Carica papaya sex chromosomes. Mol Genet Genomics 290, 1511–1522 (2015). https://doi.org/10.1007/s00438-015-1000-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-015-1000-3