Abstract
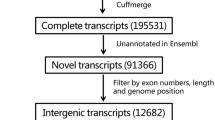
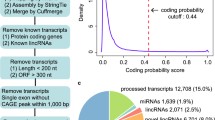
Long intergenic non-coding RNAs (lincRNAs) as a key group of non-coding RNAs have gained substantial attention. Though lincRNAs have been systematically explored in various mouse tissues and cell lines, large-scale identification of lincRNAs in mouse pre-implantation embryonic development (PED) process has not be documented previously. Therefore, it is important to identify and characterize novel lincRNAs that may be involved in PED. In this paper, we performed transcriptome assembly based on published single-cell RNA-seq data during mouse PED and identified 4,438 putative lincRNAs. Combining these with Ensembl lincRNAs, we established a reference catalog of 5,808 transcribed lincRNAs in PED. We then systematically analyzed the lincRNAs in this reference catalog and revealed that the identified novel PED lincRNAs are generally comparable with known Ensembl lincRNAs in genomic aspects. In addition, the global expression patterns can be separated by zygote first cleavage division in clustering analysis and we further identified and analyzed differentially expressed lincRNAs involved in this process. The expression of lincRNAs involved in the process is negatively correlated with promoter methylation in trend. The identified lincRNAs involved in zygote first cleavage division could have important roles in mouse early embryonic development and need further functional studies. Altogether, a novel reference catalog of mouse PED lincRNAs is provided and characterized, which would be a valuable resource for further functional analyses and may help elucidate the pre-implantation regulatory mechanism.






Similar content being viewed by others
References
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11(10):R106
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res 41(Database issue):D991–D995
Bogdanovic O, Gomez-Skarmeta JL (2014) Embryonic DNA methylation: insights from the genomics era. Brief Funct Genom 13(2):121–130
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927
Castel SE, Martienssen RA (2013) RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet 14(2):100–112
Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147(2):358–369
Deng Q, Ramskold D, Reinius B, Sandberg R (2014) Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science 343(6167):193–196
Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22(9):1775–1789
Dinger ME, Pang KC, Mercer TR, Mattick JS (2008) Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 4(11):e1000176
Esteller M (2011) Non-coding RNAs in human disease. Nat Rev Genet 12(12):861–874
Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Harrow J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM (2013) Ensembl 2013. Nucleic Acids Res 41(Database issue):D48–D55
Grote P, Wittler L, Hendrix D, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M, Herrmann BG (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24(2):206–214
Gu T, He H, Xing Y, Liu Q, Gu N, Kenkichi S, Jiang H, Wu Q (2011) Expression of non-coding RNA AB063319 derived from Rian gene during mouse development. J Mol Histol 42(2):105–112
Gu T, He H, Han Z, Zeng T, Huang Z, Liu Q, Gu N, Chen Y, Sugimoto K, Jiang H, Wu Q (2012) Expression of macro non-coding RNAs Meg8 and Irm in mouse embryonic development. Acta Histochem 114(4):392–399
Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, Rinn JL, Lander ES, Regev A (2010) Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28(5):503–510
Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477(7364):295–300
Habibi E, Brinkman AB, Arand J, Kroeze LI, Kerstens HH, Matarese F, Lepikhov K, Gut M, Brun-Heath I, Hubner NC, Benedetti R, Altucci L, Jansen JH, Walter J, Gut IG, Marks H, Stunnenberg HG (2013) Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem Cell 13(3):360–369
Hamatani T, Carter MG, Sharov AA, Ko MS (2004) Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6(1):117–131
Han Z, He H, Zhang F, Huang Z, Liu Z, Jiang H, Wu Q (2012) Spatiotemporal expression pattern of Mirg, an imprinted non-coding gene, during mouse embryogenesis. J Mol Histol 43(1):1–8
Han Z, Liu Q, Huang Z, Cui W, Tian Y, Yan W, Wu Q (2013) Expression and imprinting analysis of AK044800, a transcript from the Dlk1-Dio3 imprinted gene cluster during mouse embryogenesis. Mol Cells 35(4):285–290
He K, Zhao H, Wang Q, Pan Y (2010) A comparative genome analysis of gene expression reveals different regulatory mechanisms between mouse and human embryo pre-implantation development. Reprod Biol Endocrinol 8:41
Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C (2013) Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 9(4):e1003470
Kelley D, Rinn J (2012) Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 13(11):R107
Kung JT, Colognori D, Lee JT (2013) Long noncoding RNAs: past, present, and future. Genetics 193(3):651–669
Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338(6113):1469–1472
Lewis A, Green K, Dawson C, Redrup L, Huynh KD, Lee JT, Hemberger M, Reik W (2006) Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development 133(21):4203–4210
Liu H, Chen Y, Lv J, Zhu R, Su J, Liu X, Zhang Y, Wu Q (2013) Quantitative epigenetic co-variation in CpG islands and co-regulation of developmental genes. Sci Rep 3:2576
Liu H, Zhu R, Lv J, He H, Yang L, Huang Z, Su J, Zhang Y, Yu S, Wu Q (2014) DevMouse, the mouse developmental methylome database and analysis tools. Database (Oxford) 2014: bat084
Lorthongpanich C, Cheow LF, Balu S, Quake SR, Knowles BB, Burkholder WF, Solter D, Messerschmidt DM (2013) Single-cell DNA-methylation analysis reveals epigenetic chimerism in preimplantation embryos. Science 341(6150):1110–1112
Luo H, Sun S, Li P, Bu D, Cao H, Zhao Y (2013) Comprehensive characterization of 10,571 mouse large intergenic noncoding RNAs from whole transcriptome sequencing. PLoS ONE 8(8):e70835
Lv J, Cui W, Liu H, He H, Xiu Y, Guo J, Liu Q, Zeng T, Chen Y, Zhang Y, Wu Q (2013a) Identification and characterization of long non-coding RNAs related to mouse embryonic brain development from available transcriptomic data. PLoS ONE 8(8):e71152
Lv J, Liu H, Huang Z, Su J, He H, Xiu Y, Zhang Y, Wu Q (2013b) Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res 41(22):10044–10061
Lv J, Huang Z, Liu H, Cui W, Li B, He H, Guo J, Liu Q, Zhang Y, Wu Q (2014) Identification and characterization of long intergenic non-coding RNAs related to mouse liver development. Mol Genet Genom 289(6):1225–1235
Ma L, Bajic VB, Zhang Z (2013) On the classification of long non-coding RNAs. RNA Biol 10(6):925–933
Managadze D, Lobkovsky AE, Wolf YI, Shabalina SA, Rogozin IB, Koonin EV (2013) The vast, conserved mammalian lincRNome. PLoS Comput Biol 9(2):e1002917
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15(Spec No 1):R17–R29
McCarthy DJ, Chen Y, Smyth GK (2012) Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40(10):4288–4297
Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, Raney BJ, Pohl A, Malladi VS, Li CH, Lee BT, Learned K, Kirkup V, Hsu F, Heitner S, Harte RA, Haeussler M, Guruvadoo L, Goldman M, Giardine BM, Fujita PA, Dreszer TR, Diekhans M, Cline MS, Clawson H, Barber GP, Haussler D, Kent WJ (2013) The UCSC genome browser database: extensions and updates 2013. Nucleic Acids Res 41((Database issue)):D64–D69
Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P (2008) The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322(5908):1717–1720
Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505(7485):635–640
Pauli A, Rinn JL, Schier AF (2011) Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet 12(2):136–149
Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, Schier AF (2012) Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22(3):577–591
Ponjavic J, Ponting CP, Lunter G (2007) Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17(5):556–565
Prickett AR, Oakey RJ (2012) A survey of tissue-specific genomic imprinting in mammals. Mol Genet Genomics 287(8):621–630
Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI reference sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40((Database issue)):D130–D135
Qu Z, Adelson DL (2012) Identification and comparative analysis of ncRNAs in human, mouse and zebrafish indicate a conserved role in regulation of genes expressed in brain. PLoS ONE 7(12):e52275
Sati S, Ghosh S, Jain V, Scaria V, Sengupta S (2012) Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic Acids Res 40(20):10018–10031
Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Krasnov S, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Karsch-Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J (2012) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40((Database issue)):D13–D25
Schier AF (2007) The maternal-zygotic transition: death and birth of RNAs. Science 316(5823):406–407
Severin J, Waterhouse AM, Kawaji H, Lassmann T, van Nimwegen E, Balwierz PJ, de Hoon MJ, Hume DA, Carninci P, Hayashizaki Y, Suzuki H, Daub CO, Forrest AR (2009) FANTOM4 EdgeExpressDB: an integrated database of promoters, genes, microRNAs, expression dynamics and regulatory interactions. Genome Biol 10(4):R39
Shapiro E, Biezuner T, Linnarsson S (2013) Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14(9):618–630
Shin H, Liu T, Manrai AK, Liu XS (2009) CEAS: cis-regulatory element annotation system. Bioinformatics 25(19):2605–2606
Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D (2005) Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15(8):1034–1050
Smith ZD, Chan MM, Mikkelsen TS, Gu H, Gnirke A, Regev A, Meissner A (2012) A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484(7394):339–344
Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, Canfield T, Giste E, Johnson A, Zhang M, Balasundaram G, Byron R, Roach V, Sabo PJ, Sandstrom R, Stehling AS, Thurman RE, Weissman SM, Cayting P, Hariharan M, Lian J, Cheng Y, Landt SG, Ma Z, Wold BJ, Dekker J, Crawford GE, Keller CA, Wu W, Morrissey C, Kumar SA, Mishra T, Jain D, Byrska-Bishop M, Blankenberg D, Lajoie BR, Jain G, Sanyal A, Chen KB, Denas O, Taylor J, Blobel GA, Weiss MJ, Pimkin M, Deng W, Marinov GK, Williams BA, Fisher-Aylor KI, Desalvo G, Kiralusha A, Trout D, Amrhein H, Mortazavi A, Edsall L, McCleary D, Kuan S, Shen Y, Yue F, Ye Z, Davis CA, Zaleski C, Jha S, Xue C, Dobin A, Lin W, Fastuca M, Wang H, Guigo R, Djebali S, Lagarde J, Ryba T, Sasaki T, Malladi VS, Cline MS, Kirkup VM, Learned K, Rosenbloom KR, Kent WJ, Feingold EA, Good PJ, Pazin M, Lowdon RF, Adams LB (2012) An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13(8):418
Stower H (2013) Development: Zebrafish early methylomes. Nat Rev Genet 14(7):443
Sun K, Chen X, Jiang P, Song X, Wang H, Sun H (2013a) iSeeRNA: identification of long intergenic non-coding RNA transcripts from transcriptome sequencing data. BMC Genomics 14(Suppl 2):S7
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y (2013b) Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res 41(17):e166
Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB (2013) RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res 23(1):201–216
Tempel S (2012) Using and understanding RepeatMasker. Methods Mol Biol 859:29–51
Thierry-Mieg D, Thierry-Mieg J (2006) AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol 7 (Suppl 1): S12 11–14
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578
Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154(1):26–46
Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147(7):1537–1550
Walser CB, Lipshitz HD (2011) Transcript clearance during the maternal-to-zygotic transition. Curr Opin Genet Dev 21(4):431–443
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914
Wei Y, Su J, Liu H, Lv J, Wang F, Yan H, Wen Y, Wu Q, Zhang Y (2014) MetaImprint: an information repository of mammalian imprinted genes. Development 141(12):2516–2523
Xie D, Chen CC, Ptaszek LM, Xiao S, Cao X, Fang F, Ng HH, Lewin HA, Cowan C, Zhong S (2010) Rewirable gene regulatory networks in the preimplantation embryonic development of three mammalian species. Genome Res 20(6):804–815
Xue Z, Huang K, Cai C, Cai L, Jiang CY, Feng Y, Liu Z, Zeng Q, Cheng L, Sun YE, Liu JY, Horvath S, Fan G (2013) Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500(7464):593–597
Yan L, Yang M, Guo H, Yang L, Wu J, Li R, Liu P, Lian Y, Zheng X, Yan J, Huang J, Li M, Wu X, Wen L, Lao K, Li R, Qiao J, Tang F (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20(9):1131–1139
Zeng TB, He HJ, Zhang FW, Han ZB, Huang ZJ, Liu Q, Wu Q (2013) Expression analysis of AK003491, an imprinted noncoding RNA, during mouse development. Genes Genet Syst 88(2):127–133
Zhang Y, Liu H, Lv J, Xiao X, Zhu J, Liu X, Su J, Li X, Wu Q, Wang F, Cui Y (2011) QDMR: a quantitative method for identification of differentially methylated regions by entropy. Nucleic Acids Res 39(9):e58
Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1(3):286–298
Acknowledgments
The authors thank The Encyclopedia of DNA Elements (ENCODE) Consortium for sharing RNA-seq data. The authors also thank National Natural Science Foundation of China for funding. This work was supported by the National Natural Science Foundation of China [31171383, 31100934, 31201075, 31371478, 31371334, 61403112].
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
J. Lv, H. Liu, S. Yu, H. B. Liu, W. Cui and Y. Gao have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lv, J., Liu, H., Yu, S. et al. Identification of 4438 novel lincRNAs involved in mouse pre-implantation embryonic development. Mol Genet Genomics 290, 685–697 (2015). https://doi.org/10.1007/s00438-014-0952-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-014-0952-z