Abstract
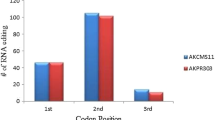
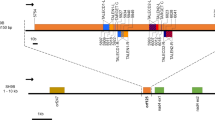
Cytoplasmic male sterility (CMS) is a maternally inherited trait resulting in failure to produce functional pollen and is widely used in the production of hybrid seed. Improper RNA editing is implicated as the molecular basis for some CMS systems. However, the mechanism of CMS in cotton is unknown. This study compared RNA editing events in eight mitochondrial genes (atp1, 4, 6, 8, 9, and cox1, 2, 3) among three lines (maintainer B, CMS A, and restorer R). These events were quantified by ultra-deep sequencing of mitochondrial transcripts and sequencing of cloned versions of these genes as cDNAs. A comparison of genomic PCR and RT-PCR products detected 72 editing sites in coding sequences in the eight genes and four partial editing sites in the 3′-untranslated region of atp6. The most frequent alteration (61.4 %) resulted in changes of hydrophilic amino acids to hydrophobic amino acids and the most common alteration was proline (P) to leucine (L) (26.7 %). In atp6, RNA editing created a stop codon from a glutamine in the genomic sequence. Statistical analysis of the frequencies of RNA editing events detected differences between mtDNA genes, but no differences between cotton cytoplasms that could account for the CMS phenotype or restoration. This study represents the first work to use next-generation sequencing to identify RNA editing positions and efficiency, and possible association with CMS and restoration in plants.



Similar content being viewed by others
References
Araya A, Domec C, Bégu D, Litvak S (1992) An in vitro system for the editing of ATP synthase subunit 9 mRNA using wheat mitochondrial extracts. Proc Natl Acad Sci USA 89:1040–1044
Bégu D, Graves PV, Domec C, Arselin G, Litvak S, Araya A (1990) RNA editing of wheat mitochondrial ATP synthase subunit 9: direct protein and cDNA sequencing. Plant Cell 2:1283–1290
Bentolila S, Elliott LE, Hanson MR (2008) Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genet 178:1693–1708
Binder S, Marchfelder A, Brennicke A, Wissinger B (1992) RNA editing in trans-splicing intron sequences of nad2 mRNAs in Oenothera. Mitochondria 267:7615–7623
Bonen L (2008) Cis- and trans-splicing of group II introns in plant mitochondria. Mitochondrion 8:26–34
Castandet B, Choury D, Begu D, Jordana X, Araya A (2010) Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res 38:7112–7121
Chapdelaine Y, Bonen L (1991) The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a f/ww-splicing model for this gene-in-pieces. Cell 65:465–472
Chase CD (2007) Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet 23:81–90
Corneille S, Lutz K, Maliga P (2000) Conservation of RNA editing between rice and maize plastids are most editing events dispensable? Mol Gen Genet 264:419–424
Covello PS, Gray MW (1989) RNA editing in plant mitochondria. Nature 341:662–666
Covello PS, Gray MW (1990) Differences in editing at homologous sites in messenger RNAs from angiosperm mitochondria. Nucleic Acids Res 18:5189–5196
Das S, Sen S, Chakraborty A, Chakraborti P, Maiti MK, Basu A, Basu D, Sen SK (2010) An unedited 1.1 kb mitochondrial orfB gene transcript in the wild abortive cytoplasmic male sterility (WA-CMS) system of Oryza sativa L. subsp. Indica. BMC Plant Biol 10:39
Fujii S, Small I (2011) The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol 191:37–47
Gallagher LJ, Betz SK, Chase CD (2002) Mitochondrial RNA editing truncates a chimeric open reading frame associated with S male-sterility in maize. Curr Genet 42:179–184
Giegé P, Brennicke A (1999) RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci USA 96:15324–15329
Grewe F, Herres S, Viehöver P, Polsakiewicz M, Weisshaar B, Knoop V (2011) A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res 39:2890–2902
Gualberto JM, Lamattina L, Bonnard G, Weil JH, Grienenberger JM (1989) RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 341:660–662
Gualberto JM, Weil JH, Grienenberger JM (1990) Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res 18:3771–3776
Handa H (2003) The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res 31:5907–5916
Hanson MR, Bentolila S (2004) Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16:S154–S169
Hanson MR, Sutton CA, Lu BW (1996) Plant organelle gene expression: altered by RNA editing. Trends Plant Sci 1:57–64
Hiesel R, Wissinger B, Schuster W, Brennicke A (1989) RNA editing in plant mitochondria. Science 246:1632–1634
Howad W, Kempken F (1997) Cell type-specific loss of atp6 RNA editing in cytoplasmic male sterile Sorghum bicolor. Proc Natl Acad Sci USA 94:11090–11095
Hu J, Wang K, Huang W, Liu G, Gao Y, Wang J, Huang Q, Ji Y, Qin X, Wan L, Zhu R, Li S, Yang D, Zhu Y (2012) The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24:109–122
Jiang W, Yang S, Yu D, Gai J (2011) A comparative study of ATPase subunit 9 (Atp9) gene between cytoplasmic male sterile line and its maintainer line in soybeans. Afr J Biotech 10:10387–10392
Knoop V, Schuster W, Wissinger B, Brennicke A (1991) Trans splicing integrates an exon of 22 nucleotides into the nad5 mRNA in higher plant mitochondria. EMBO J 10:3483–3493
Kubo T, Nishizawa S, Sugawara A, Itchoda N, Estiati A, Mikami T (2000) The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res 28:2571–2576
Kudla J, Bock R (1999) RNA editing in an untranslated region of the Ginkgo chloroplast genome. Gene 234:81–86
Kurek I, Ezra D, Begu D, Erel N, Litvak S, Breiman A (1997) Studies on the effects of nuclear background and tissue specificity on RNA editing of the mitochondrial ATP synthase subunits α, 6 and 9 in fertile and cytoplasmic malesterile (CMS) wheat. Theor Appl Genet 95:1305–1311
Lamattina L, Weil JH, Grienenberger JM (1989) RNA editing at a splicing site of NADH dehydrogenase subunit IV gene transcript in wheat mitochondria. FEBS Lett 258:79–83
Liu YG, Chen Y (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTech 43:649–656
Liu YG, Whittier RF (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25:674–681
Liu YG, Mitsukawa N, Oosumi T, Whittier RF (1995) Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 8:457–463
Mazers GR, Moyret C, Jeanteur P, Theillet C-G (1991) Direct sequencing by thermal asymmetric PCR. Nucleic Acids Res 19:4783
Mower JP, Palmer JD (2006) Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol Gen Genomics 276:285–293
Okuda K, Hammani K, Tanz SK, Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T (2010) The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. Plant J 61:339–349
Pang M, Stewart JMcD, Zhang J (2011) A mini-scale hot borate method for the isolation of total RNA from a large number of cotton tissue samples. Afr J Biotech 10:15430–15437
Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G (2010) Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucl Acids Res 38:4755–4767
Salazar RA, Pring DR, Kempken F (1991) Editing of mitochondrial atp9 transcripts from two sorghum lines. Curr Genet 20:483–486
Schnable PS, Wise RP (1998) The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci 3:175–180
Schuster W, Unseld M, Wissinger B, Brennicke A (1990) Ribosomal protein S14 transcripts are edited in Oenothera mitochondria. Nucleic Acids Res 18:229–233
Schuster W, Ternes R, Knoop V, Hiesel R, Wissinger B, Brennicke A (1991) Distribution of RNA editing sites in Oenothera mitochondrial mRNAs and rRNAs. Curr Genet 20:397–404
Sosso D, Mbelo S, Vernoud V, Gendrot G, Dedieu A, Chambrier P, Dauzat M, Heurtevin L, Guyon V, Takenaka M, Rogowsky PM (2012) PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24:676–691
Suzuki H, Yu J, Wang F, Zhang J (2013) Identification of mitochondrial DNA sequence variation and development of single nucleotide polymorphic markers for CMS-D8 in cotton. Theor Appl Genet. doi:10.1007/s00122-013-2070-4
Tan HQ, Singh J (2011) High-efficiency thermal asymmetric interlaced (HE-TAIL) PCR for amplification of Ds transposon insertion sites in Barley. J Plant Mol Biol Biotechnol 2:9–14
Terauchi R, Kahl G (2000) Rapid isolation of promoter sequences by TAIL-PCR: the 5′-flanking regions of Pal and Pgi genes from yams (Dioscorea). Mol Gen Genet 263:554–560
Unseld M, Marienfeld JR, Brandt P, Brennicke A (1997) The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924. Nat Genet 15:57–61
Wan CY, Wilkins TA (1994) A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223:7–12
Wang F, Feng CD, O’Connell MA, Stewart JMcD, Stewart JF (2010) RFLP analysis of mitochondrial DNA in two cytoplasmic male sterility systems (CMS-D2 and CMS-D8) of cotton. Euphytica 172:93–99
Wei L, Yan ZX, Ding Y (2008) Mitochondrial RNA editing of F0-ATPase subunit 9 gene (atp9) transcripts of Yunnan purple rice cytoplasmic male sterile line and its maintainer line. Acta Physiol Plant 30:657–662
Wilkins TA, Smart LB (1996) Isolation of RNA from plant tissue. In: Krieg PA (ed) A laboratory guide to RNA: isolation, analysis, and synthesis. Wiley-Liss Inc., New York, pp 21–42
Yura K, Go M (2008) Correlation between amino acid residues converted by RNA editing and functional residues in protein three-dimensional structures in plant organelles. BMC Plant Biol 8:79
Zhang J, Stewart JMcD (2000) Economical and rapid method for extracting cotton genomic DNA. J Cott Sci 4: 193-201
Zhang J, Stewart J McD (2001) Inheritance and genetic relationships of the D8 and D2-2 restorer genes for cotton cytoplasmic male sterility. Crop Sci 41:289–294
Zhang JF, Turley RB, Stewart JMcD (2008) Comparative analysis of gene expression between CMS-D8 restored plants and normal non-restoring fertile plants in cotton by differential display. Plant Cell Rep 27:553–561
Acknowledgments
The research was supported in part by New Mexico Agricultural Experiment Station. The authors thank Anthony Aragon and Jeremy Edwards for help with the Ion Torrent sequencing. Some of the experiments used the Keck-UNM Genomics Resource in the University of New Mexico Cancer Center. Parts of this work were supported by USPHS/NIH grant 1R01CA170250-01 (to SAN).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Suzuki, H., Yu, J., Ness, S.A. et al. RNA editing events in mitochondrial genes by ultra-deep sequencing methods: a comparison of cytoplasmic male sterile, fertile and restored genotypes in cotton. Mol Genet Genomics 288, 445–457 (2013). https://doi.org/10.1007/s00438-013-0764-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-013-0764-6