Abstract
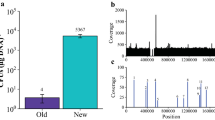
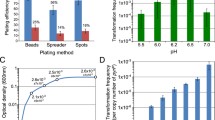
Methane is a key intermediate in the carbon cycle and biologically produced by methanogenic archaea. Most methanogens are able to conserve energy by reducing CO2 to methane using molecular hydrogen as electron donor (hydrogenotrophic methanogenesis), but several hydrogenotrophic methanogens can also use formate as electron donor for methanogenesis. Formate dehydrogenase (Fdh) oxidizes formate to CO2 and is involved in funneling reducing equivalents into the methanogenic pathway, but details on other factors relevant for formate-dependent physiology of methanogens are not available. To learn more about the factors involved in formate-dependent growth of Methanococcus maripaludis strain JJ, we used a recently developed system for random in vitro mutagenesis, which is based on a modified insect transposable element to create 2,865 chromosomal transposon mutants and screened them for impaired growth on formate. Of 12 M. maripaludis transposon-induced mutants exhibiting this phenotype, the transposon insertion sites in the chromosome were mapped. Among the genes, apparently affecting formate-dependent growth were those encoding archaeal transcription factor S, a regulator of ion transport, and carbon monoxide dehydrogenase/acetyl-CoA synthase. Interestingly, in seven of the mutants, transposons were localized in a 10.2 kb region where Fdh1, one of two Fdh isoforms in the organism, is encoded. Two transcription start sites within the 10.2 kb region could be mapped, and quantification of transcripts revealed that transposon insertion in this region diminished fdhA1 expression due to polar effects.




Similar content being viewed by others
References
Ausubel FM, Brent R, Kingston RE et al (2003) Current protocols in molecular biology. J. Wiley & Sons, Inc., New York
Bose A, Kulkarni G, Metcalf WW (2009) Regulation of putative methyl-sulphide methyltransferases in Methanosarcina acetivorans C2A. Mol Microbiol 74:227–238
Costa KC, Wong PM, Wang T et al (2010) Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc Natl Acad Sci USA 107:11050–11055
Costa KC, Yoon SH, Pan M, Burn JA, Baliga NS, Leigh JA (2013) Effects of H2 and formate on growth yield and regulation of methanogenesis in Methanococcus maripaludis. J Bacteriol 195:1456–1462
Deppenmeier U, Müller V (2008) Life close to the thermodynamic limit: how methanogenic archaea conserve energy. In: Schäfer G, Penefsky HS (eds) Bioenergetics: energy conservation and conversion. Springer, Heidelberg, pp 123–152
Ferry JG (1990) Formate dehydrogenase. FEMS Microbiol Rev 87:377–382
Fiedler S, Wirth R (1988) Transformation of bacteria with plasmid DNA by electroporation. Anal Biochem 170:38–44
Guss AM, Mukhopadhyay B, Zhang JK, Metcalf WW (2005) Genetic analysis of mch mutants in two Methanosarcina species demonstrates multiple roles for the methanopterin-dependent C-1 oxidation/reduction pathway and differences in H2 metabolism between closely related species. Mol Microbiol 55:1671–1680
Haldimann A, Wanner BL (2001) Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol 183:6384–6393
Hausner W, Lange U, Musfeldt M (2000) Transcription factor S, a cleavage induction factor of the archaeal RNA polymerase. J Biol Chem 275:12393–12399
Hendrickson EL, Kaul R, Zhou Y et al (2004) Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J Bacteriol 186:6956–6969
Hendrickson EL, Haydock AK, Moore BC, Whitman WB, Leigh JA (2007) Functionally distinct genes regulated by hydrogen limitation and growth rate in methanogenic Archaea. Proc Natl Acad Sci USA 104:8930–8934
Hildenbrand C, Stock T, Lange C, Rother M, Soppa J (2011) Genome copy numbers and gene conversion in methanogenic archaea. J Bacteriol 193:734–743
Jones WJ, Paynter MJB, Gupta R (1983) Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch Microbiol 135:91–97
Ladapo J, Whitman WB (1990) Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis. Proc Natl Acad Sci USA 87:5598–5602
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lampe DJ, Churchill ME, Robertson HM (1996) A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J 15:5470–5479
Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM (1999) Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA 96:11428–11433
Lange U, Hausner W (2004) Transcriptional fidelity and proofreading in Archaea and implications for the mechanism of TFS-induced RNA cleavage. Mol Microbiol 52:1133–1143
Leigh JA, Albers SV, Atomi H, Allers T (2011) Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol Rev 35:577–608
Lie TJ, Leigh JA (2003) A novel repressor of nif and glnA expression in the methanogenic archaeon Methanococcus maripaludis. Mol Microbiol 47:235–246
Lie TJ, Dodsworth JA, Nickle DC, Leigh JA (2007) Diverse homologues of the archaeal repressor NrpR function similarly in nitrogen regulation. FEMS Microbiol Lett 271:281–288
Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, Leigh JA (2012) Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci USA 109:15473–15478
Lupa B, Hendrickson EL, Leigh JA, Whitman WB (2008) Formate-dependent H2 production by the mesophilic methanogen Methanococcus maripaludis. Appl Environ Microbiol 74:6584–6590
Metcalf WW, Zhang JK, Shi X, Wolfe RS (1996) Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol 178:5797–5802
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nakata A, Amemura M, Shinagawa H (1984) Regulation of the phosphate regulon in Escherichia coli K-12: regulation of the negative regulatory gene phoU and identification of the gene product. J Bacteriol 159:979–985
Oza JP, Sowers KR, Perona JJ (2012) Linking energy production and protein synthesis in hydrogenotrophic methanogens. Biochemistry 51:2378–2389
Rother M, Mathes I, Lottspeich F, Böck A (2003) Inactivation of the selB gene in Methanococcus maripaludis: effect on synthesis of selenoproteins and their sulfur-containing homologs. J Bacteriol 185:107–114
Rother M, Sattler C, Stock T (2011) Studying gene regulation in methanogenic archaea. Methods Enzymol 494:91–110
Sarmiento BF, Leigh JA, Whitman WB (2011) Genetic systems for hydrogenotrophic methanogens. Methods Enzymol 494:43–73
Schlindwein C, Giordano G, Santini CL, Mandrand MA (1990) Identification and expression of the Escherichia coli fdhD and fdhE genes, which are involved in the formation of respiratory formate dehydrogenase. J Bacteriol 172:6112–6121
Self WT, Pierce R, Stadtman TC (2004) Cloning and heterologous expression of a Methanococcus vannielii gene encoding a selenium-binding protein. IUBMB Life 56:501–507
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Stadtman T (2004) Methanococcus vannielii selenium metabolism: purification and N-terminal amino acid sequences of a novel selenium-binding protein and selenocysteine lyase. IUBMB Life 56:427–431
Stock T, Rother M (2009) Selenoproteins in archaea and gram-positive bacteria. Biochim Biophys Acta 1790:1520–1532
Stock T, Selzer M, Rother M (2010) In vivo requirement of selenophosphate for selenoprotein synthesis in archaea. Mol Microbiol 75:149–160
Stock T, Selzer M, Connery S, Seyhan D, Resch A, Rother M (2011) Disruption and complementation of the selenocysteine biosynthesis pathway reveals a hierarchy of selenoprotein gene expression in the archaeon Methanococcus maripaludis. Mol Microbiol 82:734–747
Studier FW, Moffatt BA (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol 189:113–130
Tabor S, Richardson CC (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA 82:1074–1078
Thauer RK (1998) Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology 144:2377–2406
Tumbula DL, Makula RA, Whitman WB (1994) Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol Lett 121:309–314
Wanner BL (1986) Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol 191:39–58
Whitman WB, Shieh J, Sohn S, Caras DS, Premachandran U (1986) Isolation and characterisation of 22 mesophilic methanococci. Syst Appl Microbiol 7:235–240
Wood GE, Haydock AK, Leigh JA (2003) Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol 185:2548–2554
Xia Q, Hendrickson EL, Zhang Y et al (2006) Quantitative proteomics of the archaeon Methanococcus maripaludis validated by microarray analysis and real time PCR. Mol Cell Proteomics 5:868–881
Xu XM, Turanov AA, Carlson BA et al (2011) Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc Natl Acad Sci USA 107:21430–21434
Yuan J, Hohn MJ, Sherrer RL, Palioura S, Su D, Söll D (2010) A tRNA-dependent cysteine biosynthesis enzyme recognizes the selenocysteine-specific tRNA in Escherichia coli. FEBS Lett 584:2857–2861
Zhang JK, Pritchett MA, Lampe DJ, Robertson HM, Metcalf WW (2000) In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc Natl Acad Sci USA 97:9665–9670
Acknowledgments
We thank Deniz Seyhan (Frankfurt am Main, Germany) for his help in screening mini-MAR367 mutants and Marina Totrova for excellent technical assistance. The authors declare that they have no conflict of interest. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (via SFB 902) to M.R.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Klug.
Nucleotide sequence data reported are available in the GenBank database under accession number KC342234.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sattler, C., Wolf, S., Fersch, J. et al. Random mutagenesis identifies factors involved in formate-dependent growth of the methanogenic archaeon Methanococcus maripaludis . Mol Genet Genomics 288, 413–424 (2013). https://doi.org/10.1007/s00438-013-0756-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-013-0756-6