Abstract
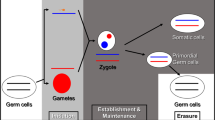
Genomic imprinting, a genetic phenomenon of non-equivalent allele expression that depends on parental origins, has been ubiquitously observed in nature. It does not only control the traits of growth and development but also may be responsible for survival traits. Based on the accelerated failure time model, we construct a general parametric model for mapping the imprinted QTL (iQTL). Within the framework of interval mapping, maximum likelihood estimation of iQTL parameters is implemented via EM algorithm. The imprinting patterns of the detected iQTL are statistically tested according to a series of null hypotheses. BIC model selection criterion is employed to choose an optimal baseline hazard function with maximum likelihood and parsimonious parameters. Simulations are used to validate the proposed mapping procedure. A published dataset from a mouse model system was used to illustrate the proposed framework. Results show that among the five commonly used survival distributions, Log-logistic distribution is the optimal baseline hazard function for mapping QTL of hyperoxic acute lung injury (HALI) survival; under the log-logistic distribution, four QTLs were identified, in which only one QTL was inherited in Mendelian fashion, whereas others were imprinted in different imprinting patterns.


Similar content being viewed by others
References
Broman KW (2003) Mapping quantitative trait loci in the case of a spike in the phenotype distribution. Genetics 163:1169–1175
Cheng JY, Tzeng S (2009) Parametric and semiparametric methods for mapping quantitative trait loci. Comput Statist Data Anal 53:1843–1849
Cheverud JM, Hager R, Roseman C, Fawcett G, Wang B, Wolf JB (2008) Genomic imprinting effects on adult body composition in mice. Proc Natl Acad Sci USA 105:4253–4258
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
Cox D, Oakes D (1984) Analysis of survival data. Chapman and Hall, London
Cui Y (2007) A statistical framework for genome-wide scanning and testing of imprinted quantitative trait loci. J Theor Biol 244:115–126
Cui Y, Lu Q, Cheverud JM, Littell RC, Wu R (2006) Model for mapping imprinted quantitative trait loci in an inbred F2 design. Genomics 87:543–551
Cui Y, Cheverud JM, Wu R (2007) A statistical model for dissecting genomic imprinting through genetic mapping. Genetica 130:227–239
de Koning DJ, Rattink AP, Harlizius B, van Arendonk JA, Brascamp EW, Groenen MA (2000) Genome-wide scan for body composition in pigs reveals important role of imprinting. Proc Natl Acad Sci USA 97:7947–7950
de Koning DJ, Bovenhuis H, van Arendonk JA (2002) On the detection of imprinted quantitative trait loci in experimental crosses of outbred species. Genetics 161:931–938
Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood from incomplet data via the EM algorithm. J R Statist Soc B 39:1–38
Diao G, Lin DY (2005) Semiparametric methods for mapping quantitative trait loci with censored data. Biometrics 61:789–798
Diao G, Lin DY, Zou F (2004) Mapping quantitative trait loci with censored observations. Genetics 168:1689–1698
Fang Y (2006) A note on QTL detecting for censored traits. Genet Sel Evol 38:221–229
Hager R, Cheverud JM, Wolf JB (2009) Relative contribution of additive, dominance, and imprinting effects to phenotypic variation in body size and growth between divergent selection lines of mice. Evolution 63:1118–1128
Hanson RL, Kobes S, Lindsay RS, Knowler WC (2001) Assessment of parent-of-origin effects in linkage analysis of quantitative traits. Am J Hum Genet 68:951–962
Jin Z, Lin DY, Wei LJ, Ying Z (2003) Rank-based inference for the accelerated failure time model. Biometrika 90:341–353
Kalbfleisch JD, Prentice RL (2002) The statistical analysis of failure time data. Wiley, New York
Kao CH, Zeng ZB, Teasdale RD (1999) Multiple interval mapping for quantitative trait loci. Genetics 152:1203–1216
Knott SA, Elsen JM, Haley CS (1996) Methods for multiple-marker mapping of quantitative trait loci in half-sib populations. Theor Appl Genet 93:71–80
Li G, Cui Y (2009) A statistical variance components framework for mapping imprinted quantitative trait loci in experimental crosses. J Prob Stat 2009:1–27
Li G, Cui Y (2010) A general statistical framework for dissecting parent-of-origin effects underlying triploid endosperm traits in flowering plants. Ann Appl Stat 3:1214–1233
Ma ZS, Bechinski EJ (2009) Accelerated failure time (AFT) modeling for the development and survival of Russian wheat aphid, Diuraphis noxia (Mordvilko). Popul Ecol 51:543–548
Mantey C, Brockmann GA, Kalm E, Reinsch N (2005) Mapping and exclusion mapping of genomic imprinting effects in mouse F2 families. J Hered 96:329–338
Moreno CR, Elsen JM, Le Roy P, Ducrocq V (2005) Interval mapping methods for detecting QTL affecting survival and time-to-event phenotypes. Genet Res 85:139–149
Pfeifer K (2000) Mechanisms of genomic imprinting. Am J Hum Genet 67:777–787
Prows DR, Hafertepen AP, Gibbons WJ Jr, Winterberg AV, Nick TG (2007a) A genetic mouse model to investigate hyperoxic acute lung injury survival. Physiol Genomics 30(3):262–270
Prows DR, Hafertepen AP, Winterberg AV, Gibbons WJ Jr, Liu C, Nick TG (2007b) Genetic analysis of hyperoxic acute lung injury survival in reciprocal intercross mice. Physiol Genomics 30(3):271–281
Schwarz G (1978) Estimating the dimension of a model. Ann Stat 6:461–464
Shete S, Amos CI (2002) Testing for genetic linkage in families by a variance-components approach in the presence of genomic imprinting. Am J Hum Genet 70:751–757
Sillanpää MJ, Arjas E (1998) Bayesian mapping of multiple quantitative trait loci from incomplete inbred line cross data. Genetics 148:1373–1388
Symons RC, Daly MJ, Fridlyand J, Speed TP, Cook WD, Gerondakis S, Harris AW, Foote SJ (2002) Multiple genetic loci modify susceptibility to plasmacytoma-related morbidity in E(mu)-v-abl transgenic mice. Proc Natl Acad Sci USA 99:11299–11304
Tuiskula-Haavisto M, de Koning DJ, Honkatukia M, Schulman NF, Maki-Tanila A, Vilkki J (2004) Quantitative trait loci with parent-of-origin effects in chicken. Genet Res 84:57–66
Wang H, Zhang YM, Li X, Masinde GL, Mohan S, Baylink DJ, Xu S (2005) Bayesian shrinkage estimation of quantitative trait loci parameters. Genetics 170:465–480
Wolf JB, Cheverud JM, Roseman C, Hager R (2008) Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genet 4:1–12
Yang R, Wang X, Wu Z, Prows DR, Lin M (2010) Bayesian model selection for characterizing genomic imprinting effects and patterns. Bioinformatics 26:235–241
Yi N (2004) A unified Markov chain Monte Carlo framework for mapping multiple quantitative trait loci. Genetics 167:967–975
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (30972077).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
Zhou, X., Fang, M., Li, J. et al. Characterization of genomic imprinting effects and patterns with parametric accelerated failure time model. Mol Genet Genomics 287, 67–75 (2012). https://doi.org/10.1007/s00438-011-0661-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-011-0661-9