Abstract
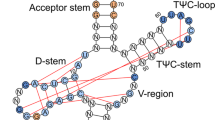
Helix 69 of 23S rRNA forms one of the major inter-subunit bridges of the 70S ribosome and interacts with A- and P-site tRNAs and translation factors. Despite the proximity of h69 to the decoding center and tRNAs, the contribution of h69 to the tRNA selection process is unclear: previous genetic analyses have shown that h69 mutations increase frameshifting and readthrough of stop codons. However, a complete deletion of h69 does not affect the selection of cognate tRNAs in vitro. To address these discrepancies, the in vivo effects of a range of single- and multi-base h69 mutations in Escherichia coli 23S rRNA on various translation errors have been determined. While a majority of the h69 mutations examined here affected readthrough of stop codons and frameshifting, the ΔA1916 single base deletion mutation uniquely influenced missense decoding. Different h69 mutants had either increased or decreased levels of stop codon readthrough. The h69 mutations that decreased UGA readthrough also decreased UGA reading by a mutant, near-cognate tRNATrp carrying a G24A substitution in the D arm, but had far less effect on UGA reading by a suppressor tRNA with a complementary anticodon. These results suggest that h69 interactions with release factors contribute significantly to termination efficiency and that interaction with the D arm of A-site tRNA is important for discrimination between cognate and near-cognate tRNAs.


Similar content being viewed by others
References
Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF (2006) Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell 23:865–874
Andersson DI, Bohman K, Isaksson LA, Kurland CG (1982) Translation rates and misreading characteristics of rpsD mutants in Escherichia coli. Mol Gen Genet 187:467–472
Asai T, Zaporojets D, Squires C, Squires CL (1999a) An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc Natl Acad Sci USA 96:1971–1976
Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL (1999b) Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol 181:3803–3809
Atkins JF, Ryce S (1974) UGA and non-triplet suppressor reading of the genetic code. Nature 249:527–530
Borovinskaya MA, Pai RD, Zhang W, Schuwirth BS, Holton JM, Hirokawa G, Kaji H, Kaji A, Cate JH (2007) Structural basis for aminoglycoside inhibition of bacterial ribosome recycling. Nat Struct Mol Biol 14:727–732
Cochella L, Green R (2005) An active role for tRNA in decoding beyond codon: anticodon pairing. Science 308:1178–1180
Cupples CG, Miller JH, Huber RE (1990) Determination of the roles of Glu-461 in beta-galactosidase (Escherichia coli) using site-specific mutagenesis. J Biol Chem 265:5512–5518
Dallas A, Noller HF (2001) Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8:855–864
Dinçbas-Renqvist V, Engström A, Mora L, Heurgué-Hamard V, Buckingham R, Ehrenberg M (2000) A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J 19:6900–6907
Ejby M, Sørensen MA, Pedersen S (2007) Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc Natl Acad Sci USA 104:19410–19415
Frank J, Verschoor A, Li Y, Zhu J, Lata RK, Radermacher M, Penczek P, Grassucci R, Agrawal RK, Srivastava S (1995) A model of the translational apparatus based on a three-dimensional reconstruction of the Escherichia coli ribosome. Biochem Cell Biol 73:757–765
Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, Chapman MS, Frank J (2003) Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113:789–801
Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J (2005) Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18:663–674
Gregory ST, Brunelli CA, Lodmell JS, O’Connor M, Dahlberg AE (1998) Genetic selection of rRNA mutations. Methods Mol Biol 77:271–281
Higuchi R, Krummel B, Saiki RK (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res 16:7351–7367
Hirabayashi N, Sato NS, Suzuki T (2006) Conserved loop sequence of helix 69 in Escherichia coli 23 S rRNA is involved in A-site tRNA binding and translational fidelity. J Biol Chem 281:17203–17211
Hirsh D (1971) Tryptophan transfer RNA as the UGA suppressor. J Mol Biol 58:439–458
Johansen SK, Maus CE, Plikaytis BB, Douthwaite S (2006) Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2′-O-methylations in 16S and 23S rRNAs. Mol Cell 23:173–182
Kipper K, Hetényi C, Sild S, Remme J, Liiv A (2009) Ribosomal intersubunit bridge B2a is involved in factor-dependent translation initiation and translational processivity. J Mol Biol 385:405–422
Klaholz BP, Pape T, Zavialov AV, Myasnikov AG, Orlova EV, Vestergaard B, Ehrenberg M, van Heel M (2003) Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature 421:90–94
Klaholz BP, Myasnikov AG, Van Heel M (2004) Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature 427:862–865
Korostelev A, Trakhanov S, Laurberg M, Noller HF (2006) Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 126:1065–1077
Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF (2008) Structural basis for translation termination on the 70S ribosome. Nature 454:852–857
Liiv A, O’Connor M (2006) Mutations in the intersubunit bridge regions of 23S rRNA. J Biol Chem 281:29850–29862
Liiv A, Karitkina D, Maivali U, Remme J (2005) Analysis of the function of E. coli 23S rRNA helix-loop 69 by mutagenesis. BMC Mol Biol 6:18
Maivali U, Remme J (2004) Definition of bases in 23S rRNA essential for ribosomal subunit association. RNA 10:600–604
Marrero P, Cabanas MJ, Modolell J (1980) Induction of translational errors (misreading) by tuberactinomycins and capreomycins. Biochem Biophys Res Commun 97:1042–1047
Matsumura K, Ito K, Kawazu Y, Mikuni O, Nakamura Y (1996) Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J Mol Biol 258:588–599
Miller JH (1991) A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Mitchell P, Osswald M, Brimacombe R (1992) Identification of intermolecular RNA cross-links at the subunit interface of the Escherichia coli ribosome. Biochemistry 31:3004–3011
Monshupanee T, Gregory ST, Douthwaite S, Chungjatupornchai W, Dahlberg AE (2008) Mutations in conserved helix 69 of 23S rRNA of Thermus thermophilus that affect capreomycin resistance but not posttranscriptional modifications. J Bacteriol 190:7754–7776
Nilsson M, Rydén-Aulin M (2003) Glutamine is incorporated at the nonsense codons UAG and UAA in a suppressor-free Escherichia coli strain. Biochim Biophys Acta 1627:1–6
O’Connor M (2007) Interaction between the ribosomal subunits: 16S rRNA suppressors of the lethal DeltaA1916 mutation in the 23S rRNA of Escherichia coli. Mol Genet Genomics 278:307–315
O’Connor M, Dahlberg AE (1995) The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J Mol Biol 254:838–847
O’Connor M, Thomas CL, Zimmermann RA, Dahlberg AE (1997) Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res 25:1185–1193
O’Connor M, Gregory ST, Dahlberg AE (2004) Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res 32:5750–5756
Raftery LA, Egan JB, Cline SW, Yarus M (1984) Defined set of cloned termination suppressors: in vivo activity of isogenetic UAG, UAA, and UGA suppressor tRNAs. J Bacteriol 158:849–859
Rawat U, Gao H, Zavialov A, Gursky R, Ehrenberg M, Frank J (2006) Interactions of the release factor RF1 with the ribosome as revealed by cryo-EM. J Mol Biol 357:1144–1153
Raychaudhuri S, Conrad J, Hall BG, Ofengand J (1998) A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4:1407–1417
Sambrook JF, Fan DP, Brenner S (1967) A strong suppressor specific for UGA. Nature 214:452–453
Schuette JC, Murphy FV 4th, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA, Ramakrishnan V, Spahn CM (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28:755–765
Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 A resolution. Science 310:827–834
Selmer M, Dunham CM, Murphy FV 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942
Stoker NG, Fairweather NF, Spratt BG (1982) Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene 18:335–341
Strigini P, Gorini L (1970) Ribosomal mutations affecting efficiency of amber suppression. J Mol Biol 47:517–530
Toth MJ, Murgola EJ, Schimmel P (1988) Evidence for a unique first position codon-anticodon mismatch in vivo. J Mol Biol 201:451–454
Vila-Sanjurjo A, Squires CL, Dahlberg AE (1999) Isolation of kasugamycin resistant mutants in the 16 S ribosomal RNA of Escherichia coli. J Mol Biol 293:1–8
Weiss RB, Dunn DM, Atkins JF, Gesteland RF (1987) Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol 52:687–693
Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V (2008) Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322:953–956
Wilson DN, Schluenzen F, Harms JM, Yoshida T, Ohkubo T, Albrecht R, Buerger J, Kobayashi Y, Fucini P (2005) X-ray crystallography study on ribosome recycling: the mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J 24:251–260
Yoshida H, Yamamoto H, Uchiumi T, Wada A (2004) RMF inactivates ribosomes by covering the peptidyl transferase centre and entrance of peptide exit tunnel. Genes Cells 9:271–278
Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 A resolution. Science 292:883–896
Acknowledgments
I am grateful to Dr. James Curran, Wake Forest University for providing the trpT mutant plasmids, to Drs. Catherine Squires and Selwyn Quan for supplying the MG1655-derived Δ7 prrn strain and to Dr. Qing Sun for constructing some of the lacZ plasmids. Thanks are due to Drs. Steven Gregory and Aivar Liiv for their comments on the manuscript. This work was supported by grants # MCB 0343942 and MCB0745025 from the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hohmann.
Rights and permissions
About this article
Cite this article
O’Connor, M. Helix 69 in 23S rRNA modulates decoding by wild type and suppressor tRNAs. Mol Genet Genomics 282, 371–380 (2009). https://doi.org/10.1007/s00438-009-0470-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-009-0470-6