Abstract
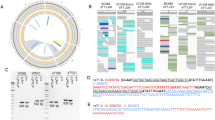
Genomic rearrangements are widely used in Caenorhabditis elegans research but many remain incompletely characterized at the physical level. We have used oligo-array comparative genomic analysis to assess the physical structure of 20 deficiencies and a single duplication of chromosome V. We find that while deletions internal to the chromosome appear simple in structure, terminal deletions are complex, containing duplications in addition to the deletion. Additionally, we confirm that transposon-induced deficiencies contain breakpoints that initiate at Tc1 elements. Finally, 13 of these deficiencies are known to suppress recombination far beyond the extent of the deletion. These deficiencies fall into two classes: strong and weak suppressors of adjacent recombination. Analysis of the deleted regions in these deficiencies reveals no common physical sites to explain the observed differences in recombination suppression. However, we find a strong correlation between the size of the rearranged chromosome and the severity of recombination suppression. Rearranged chromosomes that have a minor effect on recombination fall within 2% of normal chromosome size. Our observations highlight the use of array-based approaches for the analysis of rearranged genomes, revealing previously unidentified deficiency characteristics and addressing biologically relevant questions.





Similar content being viewed by others
References
Barrett T, Edgar R (2006) Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Methods Enzymol 411:352–369
Bessereau JL (2006) Transposons in C. elegans. WormBook 1–13
Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77(1):71–94
Clark DV, Rogalski TM, Donati LM, Baillie DL (1988) The unc-22(IV) region of Caenorhabditis elegans: genetic analysis of lethal mutations. Genetics 119(2):345–353
Clark DV, Johnsen RC, McKim KS, Baillie DL (1990) Analysis of lethal mutations induced in a mutator strain that activates transposable elements in Caenorhabditis elegans. Genome 33(1):109–114
Edgley ML, Baillie DL, Riddle DL, Rose AM (2006) Genetic balancers. WormBook 1–32. doi:10.1895/wormbook.1.89.1
Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M (2007) WormBook: the online review of Caenorhabditis elegans biology. Nucleic Acids Res 35(Database issue):D472–D475. doi:10.1093/nar/gkl894
Gresham D, Dunham MJ, Botstein D (2008) Comparing whole genomes using DNA microarrays. Nat Rev Genet 9(4):291–302. doi:10.1038/nrg2335
Hammarlund M, Davis MW, Nguyen H, Dayton D, Jorgensen EM (2005) Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics 171(3):1047–1056
Johnsen RC, Baillie DL (1988) Formaldehyde mutagenesis of the eT1 balanced region in Caenorhabditis elegans: dose-response curve and the analysis of mutational events. Mutat Res 201(1):137–147
Johnsen RC, Baillie DL (1991) Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics 129(3):735–752
Jones MR, Maydan JS, Flibotte S, Moerman DG, Baillie DL (2007) Oligonucleotide array comparative genomic hybridization (oaCGH) based characterization of genetic deficiencies as an aid to gene mapping in Caenorhabditis elegans. BMC Genomics 8(1):402
Kadandale P, Geldziler B, Hoffmann M, Singson A (2005) Use of SNPs to determine the breakpoints of complex deficiencies, facilitating gene mapping in Caenorhabditis elegans. BMC Genet 6(1):28
Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG (2007) Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res 17(3):337–347
McClintock B (1939) The behavior in successive nuclear divisions of a chromosome broken at meiosis. Proc Natl Acad Sci USA 25(8):405–416
McClintock B (1941) The Stability of broken ends of chromosomes in Zea mays. Genetics 26(2):234–282
McKim KS, Howell AM, Rose AM (1988) The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120(4):987–1001
McKim KS, Peters K, Rose AM (1993) Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134(3):749–768
Mori I, Moerman DG, Waterston RH (1988) Analysis of a mutator activity necessary for germline transposition and excision of Tc1 transposable elements in Caenorhabditis elegans. Genetics 120(2):397–407
Muller HJ (1938) The remaking of chromosomes. Collect Net 8:182–195
Muller HJ, Herskowitz IH (1954) Concerning the healing of chromosome ends produced by breakage in Drosophila melanogaster. Am Nat 88:177–208
Rogalski TM, Moerman DG, Baillie DL (1982) Essential genes and deficiencies in the unc-22 IV region of Caenorhabditis elegans. Genetics 102(4):725–736
Rose AM, Baillie DL, Curran J (1984) Meiotic pairing behavior of two free duplications of linkage group I in Caenorhabditis elegans. Mol Gen Genet 195(1–2):52–56
Rosenbluth RE, Baillie DL (1981) The genetic analysis of a reciprocal translocation, eT1(III; V), in Caenorhabditis elegans. Genetics 99(3–4):415–428
Rosenbluth RE, Cuddeford C, Baillie DL (1985) Mutagenesis in Caenorhabditis elegans. II. A spectrum of mutational events induced with 1500 r of gamma-radiation. Genetics 109(3):493–511
Rosenbluth RE, Rogalski TM, Johnsen RC, Addison LM, Baillie DL (1988) Genomic organization in Caenorhabditis elegans: deficiency mapping on linkage group V(left). Genet Res 52:105–118
Rosenbluth RE, Johnsen RC, Baillie DL (1990) Pairing for recombination in LGV of Caenorhabditis elegans: a model based on recombination in deficiency heterozygotes. Genetics 124(3):615–625
Stewart HI, O’Neil NJ, Janke DL, Franz NW, Chamberlin HM, Howell AM, Gilchrist EJ, Ha TT, Kuervers LM, Vatcher GP, Danielson JL, Baillie DL (1998) Lethal mutations defining 112 complementation groups in a 4.5 Mb sequenced region of Caenorhabditis elegans chromosome III. Mol Gen Genet 260(2–3):280–288
Wicky C, Villeneuve AM, Lauper N, Codourey L, Tobler H, Muller F (1996) Telomeric repeats (TTAGGC) n are sufficient for chromosome capping function in Caenorhabditis elegans. Proc Natl Acad Sci USA 93(17):8983–8988
Zetka MC, Rose AM (1992) The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics 131(2):321–332
Zhao Y, Lai K, Cheung I, Youds J, Tarailo M, Tarailo S, Rose A (2006) A mutational analysis of Caenorhabditis elegans in space. Mutat Res 601(1–2):19–29
Acknowledgments
This research was funded by the Canadian Space Agency and Natural Sciences and Engineering Research Council of Canada.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Hekimi.
Rights and permissions
About this article
Cite this article
Jones, M.R., Chua, S.Y., O’Neil, N.J. et al. High-resolution array comparative genomic hybridization analysis reveals unanticipated complexity of genetic deficiencies on chromosome V in Caenorhabditis elegans . Mol Genet Genomics 282, 37–46 (2009). https://doi.org/10.1007/s00438-009-0444-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-009-0444-8