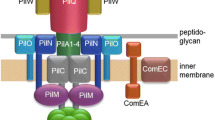
Abstract
In many type IV pili, the N-terminal amino acid of the pilin subunit is N-methylated phenylalanine. A prepilin peptidase removes the leader peptide from the precursor and methylates the amino group of the newly formed phenylalanine. PilS, the precursor of the pilin encoded by plasmid R64, is processed by the prepilin peptidase PilU, but the N-terminal amino acid of the mature pilin is a non-methylated tryptophan that is otherwise modified. To study the relationship between the structure and function of PilU, 42 missense pilU mutations were constructed by PCR and site-directed mutagenesis, and the ability of these pilU mutants to complement a pilU null mutant for mating in liquid culture was analyzed. Although practically no conjugation was noted for 21 of the mutants, the remaining 21 supported varying levels of residual plasmid transfer activity. Two mutants with aspartic acid replacements in conserved motifs exhibited no PilU activity, suggesting that the product of the pilU gene is an aspartic acid peptidase, like TcpJ, the prepilin peptidare of Vibrio cholerae. No PilS processing was detected in 21 of the mutants, but the remaining 21 exhibited varying levels of residual PilS processing. A close correlation was noted between residual PilS processing activity and conjugative transfer, suggesting that the pilU gene product possesses prepilin peptidase activity, but is unable to methylate the N-terminal tryptophan. Based on the activity of pilU-phoA and pilU-lacZ fusion genes encoding different segments of PilU, a model for the membrane topology of the protein is also proposed. Furthermore, some amino acid substitutions in the pilU portion of the pilU-phoA and pilU-lacZ fusion genes were found to alter the membrane topology of the product.






Similar content being viewed by others
References
Bally M, Filloux A, Akrim M, Ball G, Lazdunski A, Tommassen J (1992) Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol Microbiol 6:1121–1131
Bogdanov M, Heacock PN, Dowhan W (2002) A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J 21:2107–2116
Casadaban MJ, Chou J, Cohen SN (1980) In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol 143:971–980
Collyn F, Lety MA, Nair S, Escuyer V, Younes AB, Simonet M, Marceau M (2002) Yersinia pseudotuberculosis harbors a type IV pilus gene cluster that contributes to pathogenicity. Infect Immun 70:6196–6205
Duchaud E et al (2003) The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol 21:1307–1313
Frost LS (1993) Conjugative pili and pilus-specific phages. In: Clewell DB (ed) Bacterial conjugation. Plenum Press, New York, pp 189–221
Gyohda A, Furuya N, Ishiwa A, Zhu S, Komano T (2004) Structure and function of the shufflon in plasmid R64. Adv Biophys 38:183–213
Horiuchi T, Komano T (1998) Mutational analysis of plasmid R64 thin pilus prepilin: the entire prepilin sequence is required for processing by type IV prepilin peptidase. J Bacteriol 180:4613–4620
Howard SP, Critch J, Bedi A (1993) Isolation and analysis of eight exe genes and their involvement in extracellular protein secretion and outer membrane assembly in Aeromonas hydrophila. J Bacteriol 175:6695–6703
Ishiwa A, Komano T (2003) Thin pilus PilV adhesins of the plasmid R64 recognize specific structures of the lipopolysaccharide molecules of recipient cells. J Bacteriol 185:5192–5199
Johnson K, Parker ML, Lory S (1986) Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem 261:15703–15708
Kim S-R, Komano T (1992) Nucleotide sequence of the R721 shufflon. J Bacteriol 174:7053–7058
Kim S-R, Komano T (1997) The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol 179:3594–3603
Komano T (1999) Shufflons: multiple inversion systems and integrons. Annu Rev Genet 33:171–191
Komano T, Yoshida T, Narahara K, Furuya N (2000) The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol 35:1348–1359
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
LaPointe CF, Taylor RK (2000) The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem 275:1502–1510
Leyton DL, Sloan J, Hill RE, Doughty S, Hartland EL (2003) Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect Immun 71:6307–6319
Lory S, Strom MS (1997) Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa-a review. Gene 192:117–121
Manoil C (1991) Analysis of membrane protein topology using alkaline phosphatase and β-galactosidase gene fusions. Meth Cell Biol 34:61–75
Marrs CF, Schoolnik G, Koomey JM, Hardy J, Rothbard J, Falkow S (1985) Cloning and sequencing of a Moraxella bovis pilin gene. J Bacteriol 163:132–139
Mattick JS (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314
Meyer TF, Billyard E, Haas R, Storzbach S, So M (1984) Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci USA 81:6110–6114
Ogawa M, Fujitani S, Mao X, Inouye S, Komano T (1996) FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol 22:757–767
Paulsen IT, Brown MH, Dunstan SJ, Skurray RA (1995) Molecular characterization of the staphylococcal multidrug resistance export protein QacC. J Bacteriol 177:2827–2833
Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072
Pepe JC, Lory S (1998) Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa. Effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J Biol Chem 273:19120–19129
Reeves PJ, Douglas P, Salmond GP (1994) Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol Microbiol 12:445–457
Reyss I, Pugsley AP (1990) Five additional genes in the pulC–O operon of the gram-negative bacterium Klebsiella oxytoca UNF5023, which are required for pullulanase secretion. Mol Gen Genet 222:176–184
Roy CR, Isberg RR (1997) Topology of Legionalla neumophila DotA: an inner membrane protein required for replication in macrophages. Infect Immun 65:571–578
Sakai D, Komano T (2002) Genes required for plasmid R64 thin pilus biogenesis: identification and localization of products of the pilK, pilM, pilO, pilP, pilR and pilT genes. J Bacteriol 184:444–451
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Shaw CE, Taylor RK (1990) Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect Immun 58:3042–3049
Stone KD, Zhang H-Z, Carlson LK, Donnenberg MS (1996) A cluster of fourteen genes from enteropathogenic Escherichia coli is sufficient for the biogenesis of a type IV pilus. Mol Microbiol 20:325–337
Strom MS, Lory S (1993) Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol 47:565–596
Strom MS, Bergman P, Lory S (1993a) Identification of active-site cysteines in the conserved domain of PilD, the bifunctional type IV pilin leader peptidase/ N-methyltransferase of Pseudomonas aeruginosa. J Biol Chem 268:15788–15794
Strom MS, Nunn DN, Lory S (1993b) A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA 90:2404–2408
Wu SS, Kaiser D (1995) Genetic and functional evidence that Type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol 18:547–558
Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T (1998) Purification and characterization of thin pili of IncI1 plasmids ColIb-P9 and R64: formation of PilV-specific cell aggregates by type IV pili. J Bacteriol 180:2842–2848
Yoshida T, Kim S-R, Komano T (1999) Twelve pil genes are required for biogenesis of the R64 thin pilus. J Bacteriol 181:2038–2043
Zhang XL, Tsui ISM, Yip CMC, Fung AWY, Wong DKH, Dai X, Yang Y, Hackett J, Morris C (2000) Salmonella enterica serovar Typhi uses type IVB pili to enter human intestinal epithelial cells. Infect Immun 68:3067–3073
Acknowledgments
We are grateful to M. Brown for pSK4158. This work was supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by W. Goebel
Rights and permissions
About this article
Cite this article
Akahane, K., Sakai, D., Furuya, N. et al. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol Genet Genomics 273, 350–359 (2005). https://doi.org/10.1007/s00438-005-1143-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-005-1143-8