Abstract
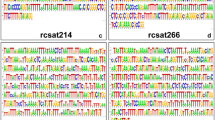
Centromeric repetitive sequences were isolated from Arabidopsis halleri ssp. gemmifera and A. lyrata ssp. kawasakiana. Two novel repeat families isolated from A. gemmifera were designated pAge1 and pAge2. These repeats are 180 bp in length and are organized in a head-to-tail manner. They are similar to the pAL1 repeats of A. thaliana and the pAa units of A. arenosa. Both A. gemmifera and A. kawasakiana possess the pAa, pAge1 and pAge2 repeat families. Sequence comparisons of different centromeric repeats revealed that these families share a highly conserved region of approximately 50 bp. Within each of the four repeat families, two or three regions showed low levels of sequence variation. The average difference in nucleotide sequence was approximately 10% within families and 30% between families, which resulted in clear distinctions between families upon phylogenetic analysis. FISH analysis revealed that the localization patterns for the pAa, pAge1 and pAge2 families were chromosome specific in A. gemmifera and A. kawasakiana. In one pair of chromosomes in A. gemmifera, and three pairs of chromosomes in A. kawasakiana, two repeat families were present. The presence of three families of centromeric repeats in A. gemmifera and A. kawasakiana indicates that the first step toward homogenization of centromeric repeats occurred at the chromosome level.





Similar content being viewed by others
References
Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G (1996) A cereal centromeric sequence. Chromosoma 105:261–268
Brandes A, Thompson H, Dean C, Heslop-Harrison JS (1997) Multiple repetitive DNA sequences in the paracentromeric regions of Arabidopsis thaliana L. Chromosome Res 5:238–246
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Cheng ZJ, Murata M (2003) A centromeric tandem repeat family originating from a part of a Ty3/gypsy -retroelement in wheat and its relatives. Genetics 164:665–672
Cheng Z, Dong F, Langdon T, Ouyang S, Buell CR, Gu M, Blattner FR, Jang J (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14:1691–1704
Choo KHA (1997) The Centromere. Oxford University Press, New York
Choo KH, Vissel B, Nagy A, Earle E, Kalitsis P (1991) A survey of the genomic distribution of alpha satellite DNA on all human chromosomes, and derivation of a new consensus sequence. Nucleic Acids Res 19:1179–1182
Copenhaver GP, Nickel K, Kuromori T, Benito MI, Kaul S, Lin X, Bevan M, Murphy G, Harris B, Parnell LD, McCombie WR, Martienssen RA, Marra M, Preuss D (1999) Genetic definition and sequence analysis of Arabidopsis centromere. Science 286:2468–2474
Csink AK, Henikoff S (1998) Something from nothing: the evolution and utility of satellite repeats. Trends Genet 14:200–204
Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J (1998) Rice ( Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci USA 95:8135–8140
Fransz P, Armstrong S, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Ward DC (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 136:867–876
Haaf T, Willard HF (1997) Chromosome-specific alpha-satellite DNA from the centromere of chimpanzee chromosome 4. Chromosoma 106:226–232
Hall SE, Kettler G, Preuss D (2003) Centromere satellites from Arabidopsis populations: maintenance of conserved and variable domains. Genome Res 13:195–205
Harrison GE, Heslop-Harrison JS (1995) Centromeric repetitive DNA sequences in the genus Brassica. Theor Appl Genet 90:157–165
Haupt W, Fischer TC, Winderl S, Fransz P, Torres-Ruiz RA (2001) The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: architecture and functional impact of chromatin. Plant J 27:285–296
Henikoff S, Ahmad K, Malik HS (2001) The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293:1098–1102
Heslop-Harrison JS (2000) Comparative genome organization in plants: from sequence and markers to chromatin and chromosomes. Plant Cell 12:617–635
Heslop-Harrison JS, Murata M, Ogura Y, Schwarzacher T, Motoyoshi F (1999) Polymorphism and genomic organization of repetitive DNA from centromeric regions of Arabidopsis thaliana. Plant Cell 11:31–42
Heslop-Harrison JS, Brandes A, Schwarzacher T (2003) Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res 11:241–253
Jiang J, Nasuda S, Dong F, Scherrer CW, Woo SS, Wing RA, Gill BS, Ward DC (1996) A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA 136:14210–14213
Kamm A, Glasso I, Schmidt T, Heslop-Harrison JS (1995) Analysis of a repetitive DNA family from Arabidopsis arenosa and relationship between Arabidopsis species. Plant Mol Biol 27:853–862
Kawabe A, Miyashita NT (1999) DNA variation in the basic chitinase locus ( ChiB) region of the wild plant Arabidopsis thaliana. Genetics 153:1445–1453
Kawabe A, Innan H, Terauchi R, Miyashita NT (1997) Nucleotide polymorphism in the acidic chitinase locus ( ChiA) region of the wild plant Arabidopsis thaliana. Mol Biol Evol 14:1303–1315
Kawabe A, Yamane K, Miyashita NT (2000) DNA polymorphism at the cytosolic phosphoglucose isomerase ( PgiC) locus of the wild plant Arabidopsis thaliana. Genetics 156:1339–1347
Kishii M, Nagaki K, Tsujimoto H (2001) A tandem repetitive sequence located in the centromeric region of common wheat ( Triticum aestivum) chromosomes. Chromosome Res 9:417–428
Koch M, Bishop J, Mitchell-Olds T (1999) Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biol 1:529–539
Koch MA, Haubold B, Mitchell-Olds T (2000) Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis , and related genera (Brassicaceae). Mol Biol Evol 17:1483–1498
Kumar S, Tamura K, Jacobsen IB, Nei M (2001) MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17:1244–1245
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2000) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 5. DNA Res 7:315–321
Kumekawa N, Hosouchi T, Tsuruoka H, Kotani H (2001) The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res 8:285–290
Laurent AM, Marcais B, Muleris M, Roizes G (1994) A rapid and simple method to isolate and characterize highly polymorphic markers from the centromeric regions of the human chromosomes. Nucleic Acids Res 22:194–199
Malik HS, Henikoff S (2001) Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157:1293–1298
Maluszynska J, Heslop-Harrison JS (1991) Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J 1:159–166
Martinez-Zapater J, Estelle MA, Somerville CR (1986) A highly repeated DNA sequence in Arabidopsis thaliana. Mol Gen Genet 204:417–423
Miyashita NT, Innan H, Terauchi R (1996) Intra- and inter specific variation in the alcohol dehydrogenase locus region of wild plants Arabis gemmifera and Arabidopsis thaliana. Mol Biol Evol 13:433–436
Miyashita NT, Kawabe A, Innan H, Terauchi R (1998) Intra- and interspecific DNA variation and codon bias of alcohol dehydrogenase ( Adh) locus in Arabis and Arabidopsis species. Mol Biol Evol 15:1420–1429
Murata M, Ogura Y, Motoyoshi F (1994) Centromeric repetitive sequence in Arabidopsis thaliana. Jpn J Genet 69:361–370
Murphy TD, Karpen GH (1998) Centromeres take flight: alpha satellite and the quest for the human centromere. Cell 93:317–320
Nagaki K, Tsujimoto H, Sasakuma T (1998) A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res 6:295–302
Nagaki K, Talbert PB, Zhong CX, Dawe RK, Henikoff S, Jiang J (2003) Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics 163:1221–1225
Pidoux AL, Allshire RC (2000) Centromeres: getting a grip of chromosomes. Curr Opin Cell Biol 12:308–319
Presting GG, Malysheva L, Fuchs J, Schubert I (1998) A TY3/GYPSY retrotransposon-like sequence localizes to the centromeric region of cereal chromosomes. Plant J 16:721–728
Richards EJ, Dawe RK (1998) Plant centromere: structure and control. Curr Opin Plant Biol 1:130–135
Round EK, Flowers SK, Richards EJ (1997) Arabidopsis thaliana centromere regions: genetic map position and repetitive DNA structure. Genome Res 7:1045–1053
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schubert I, Shi F, Fuchs J, Endo TR (1998) An efficient screening for terminal deletions and translocations of barley chromosomes added to common wheat. Plant J 14:489–495
Simoens CR, Gielen J, Van Montagu M, Inze D (1988) Characterization of highly repetitive sequence of Arabidopsis thaliana. Nucleic Acids Res 16:6753–6766
Sullivan BA, Blower MD, Karpen GH (2001) Determining centromere identity: cyclical stories and forking paths. Nature Rev Genet 2:584–596
Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S (2002) Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14:1053–1066
The Arabidopsis Genome Initiative (2000) Analysis of genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Warburton PE, Waye PE, Willard HF (1993) Nonrandom localization of recombination events in human alpha satellite repeat unit variants: implications for higher-order structural characteristics within centromeric heterochromatin. Mol Cell Biol 13:6520–6529
Warburton PE, Haaf T, Gosden J, Lawson D, Willard HF (1996) Characterization of a chromosome-specific chimpanzee alpha satellite subset: evolutionary relationship to subsets on human chromosomes. Genomics 33:220–228
Yoda K, Kitagawa K, Masumoto H, Muro Y, Okazaki T (1992) A human centromere protein, CENP-B, has a DNA-binding domain containing four potential α helices at the NH2 terminus, which is separable from dimerizing activity. J Cell Biol 119:1413–1427
Acknowledgements
The authors thank N. Goto for providing seeds. We also thank M. Murata for technical advice, suggestions and comments, and T. R. Endo for use of his facilities and for technical suggestions. We are grateful to D. Charlesworth for proofreading and suggestions. A.K. was supported by a research fellowship for young scientists awarded by the Japan Society for the Promotion of Science. This article is Contribution No. 579 from the Laboratory of Plant Genetics, Graduate School of Agriculture, Kyoto University
Author information
Authors and Affiliations
Additional information
Communicated by W. R. McCombie
Rights and permissions
About this article
Cite this article
Kawabe, A., Nasuda, S. Structure and genomic organization of centromeric repeats in Arabidopsis species. Mol Genet Genomics 272, 593–602 (2005). https://doi.org/10.1007/s00438-004-1081-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-004-1081-x