Abstract
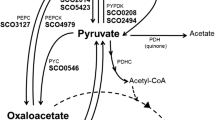
Saccharomyces kluyveri is a petite-negative yeast, which is less prone to form ethanol under aerobic conditions than is S. cerevisiae. The first reaction on the route from pyruvate to ethanol is catalysed by pyruvate decarboxylase, and the differences observed between S. kluyveri and S. cerevisiae with respect to ethanol formation under aerobic conditions could be caused by differences in the regulation of this enzyme activity. We have identified and cloned three genes encoding functional pyruvate decarboxylase enzymes ( PDC genes) from the type strain of S. kluyveri (Sk-PDC11, Sk-PDC12 and Sk-PDC13). The regulation of pyruvate decarboxylase in S. kluyveri was studied by measuring the total level of Sk-PDC mRNA and the overall enzyme activity under various growth conditions. It was found that the level of Sk-PDC mRNA was enhanced by glucose and oxygen limitation, and that the level of enzyme activity was controlled by variations in the amount of mRNA. The mRNA level and the pyruvate decarboxylase activity responded to anaerobiosis and growth on different carbon sources in essentially the same fashion as in S. cerevisiae. This indicates that the difference in ethanol formation between these two yeasts is not due to differences in the regulation of pyruvate decarboxylase(s), but rather to differences in the regulation of the TCA cycle and the respiratory machinery. However, the PDC genes of Saccharomyces/Kluyveromyces yeasts differ in their genetic organization and phylogenetic origin. While S. cerevisiae and S. kluyveri each have three PDC genes, these have apparently arisen by independent duplications and specializations in each of the two yeast lineages.




Similar content being viewed by others
References
Baburina I, Gao Y, Hu Z, Jordan F (1994) Substrate activation of brewers’ yeast pyruvate decarboxylase is abolished by mutation of cysteine 221 to serine. Biochemistry 33:5630–5635
Bianchi MM, Tizzani L, Destruelle M, Frontali L, Wésolowski-Louvel M (1996) The petite-negative yeast Kluyveromyces lactis has a single gene expressing pyruvate decarboxylase activity. Mol Microbiol 19:27–36
Destruelle M, Menghini R, Frontali L, Bianchi MM (1999) Regulation of the expression of the Kluyveromyces lactis PDC1 gene: carbon source-responsive elements and autoregulation. Yeast 15:361–370
Fauchon M, Lagniel G, Aude JC, Lombardia L, Soularue P, Petat C, Marguerie G, Sentenac A, Werner M, Labarre J (2002) Sulfur sparing in the yeast proteome in response to sulfur demand. Mol Cell 9:713–23
Flikweert MT, van der Zanden L, Janssen WMTM, Steensma HY, van Dijken JP, Pronk JT (1996) Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12:247–257
Flikweert MT, de Swaaf M, van Dijken JP, Pronk JT (1999a) Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol Lett 174:73–79
Flikweert MT, Kuyper M, van Maris AJ, Kötter P, van Dijken JP, Pronk JT (1999b) Steady-state and transient-state analysis of growth and metabolite production in a Saccharomyces cerevisiae strain with reduced pyruvate decarboxylase activity. Biotechnol Bioeng 66:42–50
Franzblau SG, Sinclair NA (1983) Induction of pyruvate decarboxylase in Candida utilis. Mycopathology 83:29–33
Gietz RD, Schiestl RH (1995) Transforming yeast with DNA. Methods Mol Cell Biol 5:255–269
Hohmann S (1991) Characterization of PDC6, a third structural gene for pyruvate decarboxylase in Saccharomyces cerevisiae. J Bacteriol 173:7963–7969
Hohmann S (1993) Characterisation of PDC2, a gene necessary for high-level expression of pyruvate decarboxylase structural genes in Saccharomyces cerevisiae. Mol Gen Genet 241:657–666
Hohmann S (1997) Pyruvate decarboxylases. In: Zimmermann FK, Entian KD (eds) Yeast sugar metabolism. Technomic Publishing AG, Basel, pp 187–211
Hohmann S, Cederberg H (1990) Autoregulation may control the expression of yeast pyruvate decarboxylase structural genes PDC1 and PDC5. Eur J Biochem 188:615–621
Holzer H (1961) Regulation of carbohydrate metabolism by enzyme competition. Cold Spring Harbor Symp Quant Biol 26:277–288
Kaliterna J, Westhuis RA, Castrillo JI, van Dijken JP, Pronk JT (1995) Coordination of sucrose uptake and respiration in the yeast Debaryomyces yamadae. Microbiology 141:1567–1574
Kellermann E, Hollenberg CP (1988) The glucose- and ethanol-dependent regulation of PDC1 from Saccharomyces cerevisiae are controlled by two distinct promoter regions. Curr Genet 14:337–344
Kiers J, Zeeman AM, Luttik MAH, Thiele C, Castrillo JI, Steensma HY, van Dijken JP, Pronk JT (1998) Regulation of alcoholic fermentation in batch and chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 14:459–469
Langkjaer RB, Nielsen ML, Daugaard PR, Liu W, Piškur J (2000) Yeast chromosomes have been significantly reshaped during their evolutionary history. J Mol Biol 304:271–288
Langkjaer RB, Cliften PF, Johnston M, Piškur J (2003) Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421:848–852
Liesen T, Hollenberg CP, Heinisch JJ (1996) ERA, a novel cis -acting element required for autoregulation and ethanol repression of PDC1 transcription in Saccharomyces cerevisiae. Mol Microbiol 21:621–632
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Møller K, Olsson L, Piškur J (2001a) Ability for anaerobic growth was not sufficient for development of the petite phenotype in Sacchromyces kluyveri. J Bacteriol 183:2485–2489
Møller K, Tidemand L, Winther JR, Olsson L, Piškur J, Nielsen J (2001b) Production of a heterologous proteinase A by Saccharomyces kluyveri. Appl Microbiol Biotechnol 57:216–219
Møller K, Christensen B, Förster J, Piškur J, Nielsen J, Olsson L (2002a) Aerobic glucose metabolism of Saccharomyces kluyveri: growth, metabolite production, and quantification of metabolic fluxes. Biotechnol Bioeng 77:186–193
Møller K, Bro C, Piškur J, Nielsen J, Olsson L (2002b) Steady-state and transient-state analyses of aerobic fermentation in Saccharomyces kluyveri. FEMS Yeast Res 2:233–244
Passoth V, Zimmermann M, Klinner U (1996) Peculiarities of the regulation of fermentation and respiration in the Crabtree-negative, xylose-fermenting yeast Pichia stipitis. Appl Biochem Biotechnol 57:201–212
Petrik M, Käppeli O, Fiechter A (1983) An expanded concept for the glucose effect in the yeast Saccharomyces uvarum: involvement of short- and long-term regulation. J Gen Microbiol 129:43–49
Phaff HJ, Miller MW, Shifrine M (1956) The taxonomy of yeasts isolated from Drosophila in the Yosemite region of California. Antonie van Leeuwenhoek 22:145–161
Piškur J, Smole S, Groth C, Petersen RF, Petersen MB (1998) Structure and genetic stability of mitochondrial genomes vary among yeasts of the genus Saccharomyces. Int J Syst Bacteriol 48:1015–1024
Pohl M (1997) Protein design on pyruvate decarboxylase (PDC) by site-directed mutagenesis. Adv Biochem Eng 58:15–43
Postma E, Verduyn C, Scheffers WA, van Dijken JP (1989) Enzymatic analysis of the Crabtree-effect in glucose-limited chemostat cultures of Saccharomyces cerevisiae. Appl Environ Microbiol 55:468–477
Prior C, Tizzani L, Fukuhara H, Wésolowski-Louvel M (1996) RAG3 gene and transcriptional regulation of the pyruvate decarboxylase gene in K. lactis. Mol Microbiol 20:765–772
Pronk JT, Wenzel TJ, Luttik MAH, Klaassen CCM, Scheffers WA, Steensma HY, van Dijken JP (1994) Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase-negative mutant of Saccharomyces cerevisiae. Microbiology 140:601–610
Pronk JT, Steensma HY, van Dijken JP (1996) Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 12:1607–1633
Remize F, Andrieu E, Dequin S (2000) Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl Environ Microbiol 66:3151–3159
Rieger M, Käppeli O, Fiechter A (1983) The role of limited respiration in the incomplete oxidation of glucose by Saccharomyces cerevisiae. J Gen Microbiol 129:653–661
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual (2nd edn). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Schmitt HD, Zimmermann FK (1982) Genetic analysis of the pyruvate decarboxylase reaction in yeast glycolysis. J Bacteriol 151:1146–1152
Schmitt HD, Ciriacy M, Zimmermann FK (1983) The synthesis of yeast pyruvate decarboxylase is regulated by large variations in the messenger RNA level. Mol Gen Genet 192:247–252
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27
Skoog K, Hahn-Hägerdal B (1990) Effect of oxygenation on xylose fermentation by Pichia stipitis. Appl Environ Microbiol 56:3389–3394
Sonnleitner B, Käppeli O (1986) Growth of Saccharomyces cerevisiae is controlled by its limited respiratory capacity: formulation and verification of a hypothesis. Biotechnol Bioeng 28:927–937
Souciet J, et al (2000) Genomic exploration of the hemiascomycetous yeasts. 1. A set of yeast species for molecular evolution studies. FEBS Lett 487:3–12
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Van de Peer Y, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Applic Biosci 10:569–570
Van Hoek P, Flikweert MT, van der Aart QJM, Steensma HY, van Dijken JP, Pronk JT (1998) Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branch point in Saccharomyces cerevisiae. Appl Environ Microbiol 64:2133–2140
Van Urk H, Schipper D, Breedveld GJ, Mak PR, Scheffers WA, van Dijken JP (1989) Localization and kinetics of pyruvate-metabolizing enzymes in relation to aerobic alcoholic fermentation in Saccharomyces cerevisiae CBS 8066 and Candida utilis CBS 621. Biochim Biophys Acta 992:78–86
Van Urk H, Voll WSL, Scheffers WA, van Dijken JP (1990) Transient-state analysis of metabolic fluxes in Crabtree-positive and Crabtree-negative yeasts. Appl Environ Microbiol 56:281–287
Venturin C, Boze H, Moulin G, Galzy P (1995a) Glucose metabolism, enzymic analysis and product formation in chemostat culture of Hanseniaspora uvarum. Yeast 11:327–336
Venturin C, Boze H, Moulin G, Galzy P (1995b) Influence of oxygen limitation on glucose metabolism in Hanseniaspora uvarum K5 grown in chemostat. Biotechnol Lett 17:537–542
Verduyn C, Postma E, Scheffers A, van Dijken JP (1992) Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517
Westhuis RA, Visser W, Pronk JT, Sheffers WA, van Dijken JP (1994) Effects of oxygen limitation on sugar metabolism in yeasts: a continuous-culture study of the Kluyver effect. Microbiology 140:703–715
Wolfe KH, Shields DC (1997) Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387:708–713
Zeeman AM, Kuyper M, Pronk JT, van Dijken JP, Steensma HY (2000) Regulation of pyruvate metabolism in chemostat cultures of Kluyveromyces lactis CBS 2359. Yeast 16:611–620
Acknowledgements
Kasper Møller was financed by a grant (No. 9900657) from the Danish Technical Research Council. Tina Johansen, Maibritt Pedersen and Jeanne Hvidtfeldt are acknowledged for their excellent assistance with the experiments. The work has been carried out in compliance with the current laws governing genetic experimentation in Denmark
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by C. P. Hollenberg
Rights and permissions
About this article
Cite this article
Møller, K., Langkjaer, R.B., Nielsen, J. et al. Pyruvate decarboxylases from the petite-negative yeast Saccharomyces kluyveri . Mol Genet Genomics 270, 558–568 (2004). https://doi.org/10.1007/s00438-003-0950-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0950-z