Abstract
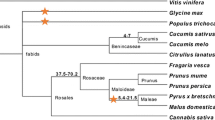
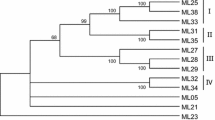
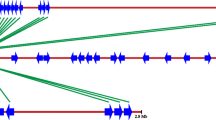
Eleven distinct families of resistance gene analogs (RGAs) with the characteristic nucleotide-binding sequence (NBS) were identified in two wild apple species, Malus prunifolia and M. baccata, and two cultivated apple cultivars, M. domestica cv. Fuji and M. domestica cv. Hong-ok, using PCR approaches with degenerate primers based on two conserved motifs of known NBS-LRR resistance genes. These RGA families were found to be represented in all the apple species tested, including wild and cultivated species. However, their sequences are very divergent from each other. Furthermore, the low level of recombination detected within their RGA families supports the idea that the evolution of NBS-encoding sequences in apple species involves the gradual accumulation of mutations. Despite the high diversity of the RGA families found in all apple species, the apparent lack of differentiation between wild and cultivated forms suggests that other factors, such as the capacity to tolerate pathogens, might play an important role in the survival of wild-type species.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Asamizu E, Sato S, Kaneko T, Nakamura Y, Kotan I, Miyajima N, Tabata S (1998) Structural analysis of Arabidopsis thaliana chromosome 5. VIII. Sequence features of the regions of 1,081,958 bp covered by seventeen physically assigned P1 and TAC clones. DNA Res 5:379–391
Bent AF (1996) Plant disease resistance gene: function meets structure. Plant Cell 8:1757–1771
Boyes DC, Nam J, Dangl JL (1998) The Arabidopsis thaliana R PM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95:15849–15854
Cooley MB, Pathirana S, Wu HJ, Kachroo P, Klessig DF (2000) Members of the Arabidopsis HRT/RPP8family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12:663–676
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
Felsenstein J (1985) Confidence limits on phylogenesis: an approach using the bootstrap. Evolution 39:783–791
Gassmann W, Hinsch ME, Staskawicz BJ (1999) The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J 20:265–277
Hammond-Kosack KE, Jones JDG (1997) Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol 48:575–607
Hehl R, Faurie E, Hesselbach J, Salamini F, Whitham S, Baker B, Gebhardt C (1999) TMV resistance gene N homologues are linked to Synchythium endobioticum resistance in potato. Theor Appl Genet 98:379–386
Kanazin V, Marek LF, Shoemaker RC (1996) Resistance gene analogs are conserved and clustered in soybean. Proc Natl Acad Sci USA 93:11746–11750
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide seqeunce. J Mol Biol 16:111–120
Kover P, Schaal B (2002) Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc Natl Acad Sci USA 99:11270–11274
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA: Molecular Evolutionary Genetics Analysis (Version 2.1). Pennsylvania State University, University Park
Leister D, Ballvora A, Salamini F, Gebhardt C (1996) A PCR-based approach for isolating pathogen resistance genes from potato with potential for wide application in plants. Nat Genet 14:421
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Noel L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at the RPP5complex disease resistance locus of Arabidopsis. Plant Cell 11:2099–2112
Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease-resistance gene homologues in coffee trees ( Coffea L.). Mol. Genet. Genomics 265:654–662
Pan Q, Liu YS, Budai-Hadrian O, M S, Carmel-Goren L, Zamir D, Fluhr R (2000) Comparative genetics of nucleotide binding site-leucine rich repeat resistance gene homologues in the genomes of two dicotyledons: tomato and Arabidopsis. Genetics 155:309–322
Posada D (2002) Evaluation of methods for detecting recombination from DNA sequences: Empirical data. Mol Biol Evol 19:708–717
Richter TE, Ronald PC (2000) The evolution of disease resistance genes. Plant Mol Biol 42:195–204
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Sawyer S (1989) Statistical tests for detecting gene conversion. Mol Biol Evol 6:526–538
Shen KA, Meyers BC, Nurul Islam-Faridi M, Chin DB, Stelly DM, Michelmore RW (1998) Resistance gene candidates identified by PCR with degenerate oligonucleotide primers map to clusters of resistance genes in lettuce. Mol Plant Microbe Interact 11:815–823
Strauss SY, Agrawal AA (1999) The ecology and evolution of plant tolerance to herbivory. Trends Ecol Evol 14:179–185
Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ (1999) Expression of the Bs2pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96:14153–14158
Van der Biezen E, Freddie C, K K, JE P, Jones J (2002) Arabidopsis RPP4is a member of the RPP5multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29:439–451
Vinatzer BA, Patocchi A, Gianfranceschi L, Tartarini S, Zhang HB, Gessler C, Sansavini S (2001) Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol Plant Microbe Interact 14:508–515
Whitham S, Dinesh-Kumar SP, Choi D, Hehl R, Corr D, Baker B (1994) The product of the tobacco mosaic virus resistance gene N: similarity to Toll and the interleukin-1 receptor. Cell 78:1101–1115
Yu YG, Buss GR, Maroof MA (1996) Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc Natl Acad Sci USA 93:11751–11756
Acknowledgements
We are grateful to Dr. Y. E. Shin (National Horticultural Research Institute, Korea) for leaves collected from apple trees. This work was supported in part by a special grant research program of the Ministry of Agriculture and Forestry of Korea (1997), by a grant from the Brain Korea 21 project (2001), and by a grant from the 21st frontier R&D program (CG124).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by R. Hagemann
Rights and permissions
About this article
Cite this article
Lee, SY., Seo, JS., Rodriguez-Lanetty, M. et al. Comparative analysis of superfamilies of NBS-encoding disease resistance gene analogs in cultivated and wild apple species. Mol Gen Genomics 269, 101–108 (2003). https://doi.org/10.1007/s00438-003-0816-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-003-0816-4