Abstract
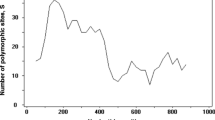
Plasmodium cynomolgi is a simian malaria parasite that has been increasingly infecting humans. It is naturally present in the long-tailed and pig-tailed macaques in Southeast Asia. The P. cynomolgi Duffy binding protein 1 region II [PcDBP1(II)] plays an essential role in the invasion of the parasite into host erythrocytes. This study investigated the genetic polymorphism, natural selection and haplotype clustering of PcDBP1(II) from wild macaque isolates in Peninsular Malaysia. The genomic DNA of 50 P. cynomolgi isolates was extracted from the macaque blood samples. Their PcDBP1(II) gene was amplified using a semi-nested PCR, cloned into a plasmid vector and subsequently sequenced. The polymorphism, natural selection and haplotypes of PcDBP1(II) were analysed using MEGA X and DnaSP ver.6.12.03 programmes. The analyses revealed high genetic polymorphism of PcDBP1(II) (π = 0.026 ± 0.004; Hd = 0.996 ± 0.001), and it was under purifying (negative) selection. A total of 106 haplotypes of PcDBP1(II) were identified. Phylogenetic and haplotype analyses revealed two groups of PcDBP1(II). Amino acid length polymorphism was observed between the groups, which may lead to possible phenotypic difference between them.





Similar content being viewed by others
Data availability
All datasets are presented in the manuscript. Nucleotide sequences generated in the study are deposited into GenBank.
References
Ahmed MA, Chu KB, Vythilingam I, Quan FS (2018) Within-population genetic diversity and population structure of Plasmodium knowlesi merozoite surface protein 1 gene from geographically distinct regions of Malaysia and Thailand. Malar J 17:442. https://doi.org/10.1186/s12936-018-2583-z
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Chin AZ, Maluda MCM, Jelip J et al (2020) Malaria elimination in Malaysia and the rising threat of Plasmodium knowlesi. J Physiol Anthropol 39:36. https://doi.org/10.1186/s40101-020-00247-5
Coatney GR (1971) The primate malarias. US National Institute of Allergy and Infectious Diseases, USA
Contacos PG, Elder HA, Coatney GR et al (1962) Man to man transfer of two strains of Plasmodium cynomolgi by mosquito bite. Am J Trop Med Hyg 11:186–193. https://doi.org/10.4269/ajtmh.1962.11.186
Daniels C (1908) Animal parasites in man and some of the lower animals in Malaya. Stud Inst Med Res FMS 3:1–13
Divis PC, Lin LC, Rovie-Ryan JJ et al (2017) Three divergent subpopulations of the malaria parasite Plasmodium knowlesi. Emerg Infect Dis 23:616–624. https://doi.org/10.3201/eid2304.161738
Doum D, Mclver DJ, Hustedt J et al (2023) An active and targeted survey reveals asymptomatic malaria infections among high-risk populations in Mondulkiri. Cambodia Malar J 22:193. https://doi.org/10.1186/s12936-023-04630-2
Fong MY, Rashdi SA, Yusof R et al (2015) Distinct genetic difference between the Duffy binding protein (PkDBPαII) of Plasmodium knowlesi clinical isolates from North Borneo and Peninsular Malaysia. Malar J 14:91. https://doi.org/10.1186/s12936-015-0610-x
Fong MY, Rashdi SA, Yusof R et al (2016) Genetic diversity, natural selection and haplotype grouping of Plasmodium knowlesi gamma protein region II (PkγRII): comparison with the Duffy binding protein (PkDBPαRII). PLoS ONE 11:e0155627. https://doi.org/10.1371/journal.pone.0155627
Fong MY, Lau YL, Jelip J et al (2019) Genetic characterisation of the erythrocyte-binding protein (PkβII) of Plasmodium knowlesi isolates from Malaysia. J Genet 98:64. https://doi.org/10.1007/s12041-019-1109-y
Gaur D, Mayer DG, Miller LH (2004) Parasite ligand–host receptor interactions during invasion of erythrocytes by Plasmodium merozoites. Int J Parasitol 34:1413–1429. https://doi.org/10.1016/j.ijpara.2004.10.010
Ju HL, Kang JM, Moon SU et al (2012) Genetic polymorphism and natural selection of Duffy binding protein of Plasmodium vivax Myanmar isolates. Malar J 11:60. https://doi.org/10.1186/1475-2875-11-60
Kojom Foko LP, Kumar A, Hawadak J et al (2023) Plasmodium cynomolgi in humans: current knowledge and future directions of an emerging zoonotic malaria parasite. Infection 51:623–640. https://doi.org/10.1007/s15010-022-01952-2
Kosaisavee V, Suwanarusk R, Chua ACY et al (2017) Strict tropism for CD71(+)/CD234(+) human reticulocytes limits the zoonotic potential of Plasmodium cynomolgi. Blood 130:1357–1363. https://doi.org/10.1182/blood-2017-02-764787
Krotoski WA, Bray RS, Garnham PC et al (1982) Observations on early and late post-sporozoite tissue stages in primate malaria. II. The hypnozoite of Plasmodium cynomolgi bastianellii from 3 to 105 days after infection, and detection of 36- to 40-hour pre-erythrocytic forms. Am J Trop Med Hyg 31:211–225. https://doi.org/10.4269/ajtmh.1982.31.211
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Latif ENM, Shahari S, Amir A et al (2022) Genetic diversity of Duffy binding protein 2 region II of Plasmodium cynomolgi from wild macaques in Peninsular Malaysia. Trop Biomed 39:66–72
Lee KS, Divis PC, Zakaria SK et al (2011) Plasmodium knowlesi: reservoir hosts and tracking the emergence in humans and macaques. PLoS Pathog 7:e1002015. https://doi.org/10.1371/journal.ppat.1002015
Mulligan H (1935) Descriptions of two species of monkey Plasmodium isolated from Silenus irus. Archir Fur Protistenkunde 84:285–314
Okenu D, Malhotra P, Lalitha PV et al (1997) Cloning and sequence analysis of a gene encoding an erythrocyte binding protein from Plasmodium cynomolgi. Mol Biochem Parasitol 89:301–306. https://doi.org/10.1016/s0166-6851(97)00118-7
Pacheco MA, Elango AP, Rahman AA et al (2012) Evidence of purifying selection on merozoite surface protein 8 (MSP8) and 10 (MSP10) in Plasmodium spp. Infect Genet Evol 12:978–986. https://doi.org/10.1016/j.meegid.2012.02.009
Pacheco MA, Reid MJ, Schillaci MA et al (2012) The origin of malarial parasites in orangutans. PLoS ONE 7:e34990. https://doi.org/10.1371/journal.pone.0034990
Putaporntip C, Kuamsab N, Jongwutiwes S (2016) Sequence diversity and positive selection at the Duffy-binding protein genes of Plasmodium knowlesi and P. cynomolgi: analysis of the complete coding sequences of Thai isolates. Infect Genet Evol 44:367–375. https://doi.org/10.1016/j.meegid.2016.07.040
Putaporntip C, Kuamsab N, Pattanawong U et al (2021) Plasmodium cynomolgi co-infections among symptomatic malaria patients, Thailand. Emerg Infect Dis 27:590–593. https://doi.org/10.3201/eid2702.191660
Raja TN, Hu TH, Kadir KA et al (2020) Naturally acquired human Plasmodium cynomolgi and P. knowlesi infections. Malaysian Borneo Emerg Infect Dis 26:1801–1809. https://doi.org/10.3201/eid2608.200343
Ranjan A, Chitnis CE (1999) Mapping regions containing binding residues within functional domains of Plasmodium vivax and Plasmodium knowlesi erythrocyte-binding proteins. Proc Natl Acad Sci USA 96:14067–14072. https://doi.org/10.1073/pnas.96.24.14067
Rice BL, Acosta MM, Pacheco MA et al (2014) The origin and diversification of the merozoite surface protein 3 (msp3) multi-gene family in Plasmodium vivax and related parasites. Mol Phylogenet Evol 78:172–184. https://doi.org/10.1016/j.ympev.2014.05.013
Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC et al (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Saat F, Nyunt MH, Abdullah NR et al (2017) Genetic diversity of Plasmodium vivax Duffy binding protein (PvDBP) gene in Sabah, Malaysia. Southeast Asian J Trop Med Public Health 48:737–748
Sai-Ngam P, Pidtana K, Suida P et al (2022) Case series of three malaria patients from Thailand infected with the simian parasite. Plasmodium Cynomolgi Malar J 21:142. https://doi.org/10.1186/s12936-022-04167-w
Sutton PL, Luo Z, Divis PC et al (2016) Characterizing the genetic diversity of the monkey malaria parasite Plasmodium cynomolgi. Infect Genet Evol 40:243–252. https://doi.org/10.1016/j.meegid.2016.03.009
Ta TH, Hisam S, Lanza M et al (2014) First case of a naturally acquired human infection with Plasmodium cynomolgi. Malar J 13:68. https://doi.org/10.1186/1475-2875-13-68
Tachibana S, Sullivan SA, Kawai S et al (2012) Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet 44:1051–1055. https://doi.org/10.1038/ng.2375
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. https://doi.org/10.1093/genetics/123.3.585
Warren M, Skinner J, Guinn E (1966) Biology of the simian malarias of Southeast Asia. I. Host cell preferences of young trophozoites of four species of Plasmodium. J Parasitol 52:14–16
Yap NJ, Hossain H, Nada-Raja T et al (2021) Natural human infections with Plasmodium cynomolgi, P. inui, and 4 other simian malaria parasites. Malaysia Emerg Infect Dis 27:2187–2191. https://doi.org/10.3201/eid2708.204502
Yusuf NM, Zulkefli J, Jiram AI et al (2022) Plasmodium spp. in macaques, Macaca fascicularis, in Malaysia, and their potential role in zoonotic malaria transmission. Parasite 29(29):32. https://doi.org/10.1051/parasite/2022032
Acknowledgements
The authors thank the Department of Wildlife and National Parks Peninsular Malaysia for their help in field macaque sampling (Permit Number: W-00256-15-19).
Funding
This study was funded by the Fundamental Research Grant Scheme (FRGS) of the Ministry of Higher Education Malaysia (Grant Number: FRGS/1/2020/SKK0/UM/01/3), awarded to Fong MY.
Author information
Authors and Affiliations
Contributions
Investigation, data curation and formal analysis: Latif ENM, Noordin NR, Shahari S; conceptualization, methodology and supervision: Fong MY, Lau YL, Amir A, Cheong FW; resources: Shahari S, Abdullah ML, Lau YL, Fong MY; writing – original draft and visualization: Latif ENM; writing – review & editing: Noordin NR, Shahari S, Amir A, Cheong FW, Lau YL, Abdullah ML, Fong MY; funding acquisition and project administration: Fong MY. The first draft of the manuscript was written by Latif ENM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This study was approved by the University of Malaya Institutional Animal Care and Use Committee (M/06122019/25022019–01/R), and the macaque blood sampling protocol was approved by UC Davis IACUC (Protocol No. 16048). All methods were performed in accordance with relevant guidelines and regulations.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Alexander Maier
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
436_2024_8125_MOESM2_ESM.tif
Supplementary file2 Supplementary data 2. Multiple alignment of deduced amino acid sequences of the 106 haplotypes corresponding to the amino acid sequences of the reference of Strain B (XM_004221494) and Berok-C2-lab adapted (OR198184). Polymorphic amino acids are listed for each haplotype. Monomorphic, dimorphic, trimorphic and tetramorphic amino acid changes are marked in yellow, blue, green, and red shadings, respectively. Numbers in the top row indicate amino acid positions. The total number of sequences for each haplotype is listed in the panel on the right. (TIF 1069 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Latif, E.N.M., Noordin, N.R., Shahari, S. et al. Genetic polymorphism and clustering of the Plasmodium cynomolgi Duffy binding protein 1 region II of recent macaque isolates from Peninsular Malaysia. Parasitol Res 123, 105 (2024). https://doi.org/10.1007/s00436-024-08125-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00436-024-08125-0