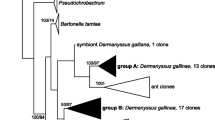
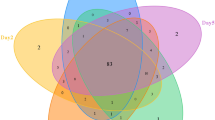
Abstract
The hematophagous arthropod, Dermanyssus gallinae (Poultry red mite, PRM) can cause remarkable economic losses in the poultry industry across the globe. Although overall composition of endosymbiotic bacteria has been shown in previous studies, how farm habitats influence the microbiome remains unclear. In the present study, we compared the bacterial communities of D. gallinae populations collected from the cage and free-range farms using next-generation sequences targeting the V3-V4 hypervariable region of the 16S rRNA gene. The QIIME2 pipeline was followed in bioinformatic analyses. Proteobacteria represented a great majority of the total bacterial community of D. gallinae from both farming systems. More specifically, Bartonella-like bacteria (40.8%) and Candidatus Cardinium (21.5%) were found to be predominant genera in free-range and cage rearing systems, respectively. However, the microbiome variation based on farming systems was not statistically significant. In addition, the presence of the five common endosymbiotic bacteria (Wolbachia, Cardinium, Rickettsiella, Spiroplasma, and Schineria) was screened in different developmental stages of D. gallinae. Cardinium was detected in all developmental stages of D. gallinae. On the other hand, Wolbachia and Rickettsiella were only found in adults/nymphs, but neither in the eggs nor larvae. To our knowledge, this study provides the first microbiome comparison at genus-level in D. gallinae populations collected from different farm habitats and will contribute to the knowledge of the biology of D. gallinae.




Similar content being viewed by others
Data availability
All relevant data are within the manuscript.
References
Akman Gündüz E, Douglas AE (2009) Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci 276(1658):987–991. https://doi.org/10.1098/rspb.2008.1476
Baumann P (2005) Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol 59:155–189. https://doi.org/10.1146/annurev.micro.59.030804.121041
Brownlie JC, Johnson KN (2009) Symbiont-mediated protection in insect hosts. Trends Microbiol 17(8):348–354. https://doi.org/10.1016/j.tim.2009.05.005
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Chauve C (1998) The poultry red mite, Dermanyssus gallinae (De Geer, 1778): current situation and future prospects for control. Vet Parasitol 79:239–245. https://doi.org/10.1016/S0304-4017(98)00167-8
Cui Y, Wang Q, Liu S, Sun R, Zhou Y, Li Y (2017) Age-related variations in intestinal microflora of free-range and caged hens. Front Microbiol 8:1310. https://doi.org/10.3389/fmicb.2017.01310
De Luna CJ, Valiente Moro C, Guy JH, Zenner L, Sparagano OA (2009) Endosymbiotic bacteria living inside the poultry red mite (Dermanyssus gallinae). Exp Appl Acarol 48(1–2):105–113. https://doi.org/10.1007/s10493-008-9230-2
Di Palma A, Giangaspero A, Cafiero MA, Germinara GS (2012) A gallery of the key characters to ease identification of Dermanyssus gallinae (Acari: Gamasida: Dermanyssidae) and allow differentiation from Ornithonyssus sylviarum (Acari: Gamasida: Macronyssidae). Parasit Vectors 5(1):1–10. https://doi.org/10.1186/1756-3305-5-104
Douglas AE (2016) How multi-partner endosymbioses function. Nat Rev Microbiol 14(12):731–743. https://doi.org/10.1038/nrmicro.2016.151
Duron O, Noël V, McCoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, Vavre F, Zenner L, Jourdain E, Durand P, Arnathau C, Renaud F, Trape JF, Chevillon C 2015 The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS pathogens 11(5):e1004892. https://doi.org/10.1371/journal.ppat.1004892
Enigl M, Schausberger P (2007) Incidence of the endosymbionts Wolbachia, Cardinium and Spiroplasma in phytoseiid mites and associated prey. Exp Appl Acarol 42(2):75–85. https://doi.org/10.1007/s10493-007-9080-3
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fukatsu T, Nikoh N (2000) Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera). AEM 66(2):643–650. https://doi.org/10.1128/AEM.66.2.643-650.2000
George DR, Finn RD, Graham KM, Mul MF, Maurer V, Valiente Moro C, Sparagano OA (2015) Should the poultry red mite Dermanyssus gallinae be of wider concern for veterinary and medical science? Parasit Vectors 8(1):1–10. https://doi.org/10.1186/s13071-015-0768-7
Goncalves LR, Harrus S, Herrera HM, Gutierrez R, Pedrassani D, Nantes WAG, Santos FM, Porfírio GEO, Barreto WTG, Macedo GG, Assis WO, Campos JBV, Silva TMV, Biolchi J, Sousa KCM, Nachum-Biala Y, Barros-Battesti DM, Machado RZ, Andre MR (2020) Low occurrence of Bartonella in synanthropic mammals and associated ectoparasites in peri-urban areas from Central-Western and Southern Brazil. Acta Trop 207:105513. https://doi.org/10.1016/j.actatropica.2020.105513
Gotoh T, Noda H, Ito S (2007) Cardinium symbionts cause cytoplasmic incompatibility in spider mites. Heredity 98(1):13–20
Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, Tzuri G, Horowitz AR, Belausov E, Mozes-Daube N, Kontsedalov S, Gershon M, Gal S, Katzir N, Zchori-Fein E (2006) Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl Environ Microbiol 72(5):3646–3652. https://doi.org/10.1128/AEM.72.5.3646-3652.2006
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH (2008) How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett 281(2):215–220. https://doi.org/10.1111/j.1574-6968.2008.01110.x
Hubert J, Erban T, Kopecky J, Sopko B, Nesvorna M, Lichovnikova M, Schicth S, Strube C, Sparagano O (2017) Comparison of microbiomes between red poultry mite populations (Dermanyssus gallinae): predominance of Bartonella-like bacteria. Microb Ecol 74(4):947–960. https://doi.org/10.1007/s00248-017-0993-z
Itoh H, Tago K, Hayatsu M, Kikuchi Y (2018) Detoxifying symbiosis: microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat Prod Rep 35(5):434–454. https://doi.org/10.1039/C7NP00051K
Iturbe-Ormaetxe I, Walker T, O’Neill SL (2011) Wolbachia and the biological control of mosquito-borne disease. EMBO Rep 12(6):508–518. https://doi.org/10.1038/embor.2011.84
Jang S, Kikuchi Y (2020) Impact of the insect gut microbiota on ecology, evolution, and industry. Curr Opin Insect Sci 41:33–39. https://doi.org/10.1016/j.cois.2020.06.004
Jeyaprakash A, Hoy MA (2000) Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol 9(4):393–405. https://doi.org/10.1046/j.1365-2583.2000.00203.x
Kageyama D, Narita S, Watanabe M (2012) Insect sex determination manipulated by their endosymbionts: incidences, mechanisms and implications. Insects 3(1):161–199. https://doi.org/10.3390/insects3010161
Koç N, İnak E, Nalbantoğlu S, Alpkent YN, Dermauw W, Van Leeuwen T (2022) Biochemical and molecular mechanisms of acaricide resistance in Dermanyssus gallinae populations from Turkey. Pestic Biochem Physiol 180:104985. https://doi.org/10.1016/j.pestbp.2021.104985
Koç N, Nalbantoğlu S (2021) Evaluation of in-house factors affecting the population distribution of Dermanyssus gallinae in cage and backyard rearing systems by using a modified monitoring method. Exp Appl Acarol 84(3):529–541. https://doi.org/10.1007/s10493-021-00638-y
Mattie DR (2005) Perchlorate. In: Wexler P, Anderson BD, Gad SC, Hakkinen PB, Kamrin M, De Peyster A, Locey B, Pope C, Mehendale HM, Shugart LR (eds) Encyclopedia of Toxicology, 2nd edn. Academic Press, New York
Montenegro H, Souza WN, Da Silva LD, Klaczko LB (2000) Male-killing selfish cytoplasmic element causes sex-ratio distortion in Drosophila melanogaster. Heredity 85(5):465–470. https://doi.org/10.1046/j.1365-2540.2000.00785.x
Moran NA, McCutcheon JP, Nakabachi A (2008) Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet 42:165–190. https://doi.org/10.1146/annurev.genet.41.110306.130119
Oliveros JC (2007) VENNY. An interactive tool for comparing lists with Venn’s diagrams. https://bioinfogp.cnb.csic.es/tools/venny/index.html. Accessed 30 July 2022.
Osaka R, Ichizono T, Kageyama D, Nomura M, Watada M (2013) Natural variation in population densities and vertical transmission rates of a Spiroplasma endosymbiont in Drosophila hydei. Symbiosis 60(2):73–78. https://doi.org/10.1007/s13199-013-0242-2
Pina T, Sabater-Muñoz B, Cabedo-López M, Cruz-Miralles J, Jaques JA, Hurtado-Ruiz MA (2020) Molecular characterization of Cardinium, Rickettsia, Spiroplasma and Wolbachia in mite species from citrus orchards. Exp Appl Acarol 81(3):335–355. https://doi.org/10.1007/s10493-020-00508-z
Price DR, Bartley K, Blake DP, Karp-Tatham E, Nunn F, Burgess ST, Nisbet AJ (2021) A Rickettsiella endosymbiont is a potential source of essential B-vitamins for the Poultry Red Mite, Dermanyssus gallinae. Front Microbiol 12:2383. https://doi.org/10.3389/fmicb.2021.695346
Roy L, Giangaspero A, Sleeckx N, Øines Ø (2021) Who is Dermanyssus gallinae? Genetic structure of populations and critical synthesis of the current knowledge. Front Vet Sci 8:650546. https://doi.org/10.3389/fvets.2021.650546
Schiavone A, Pugliese N, Otranto D, Samarelli R, Circella E, De Virgilio C, Camarda A (2022) Dermanyssus gallinae: the long journey of the poultry red mite to become a vector. Parasit Vectors 15(1):1–8. https://doi.org/10.1186/s13071-021-05142-1
Sigognault Flochlay A, Thomas E, Sparagano O (2017) Poultry red mite (Dermanyssus gallinae) infestation: a broad impact parasitological disease that still remains a significant challenge for the egg-laying industry in Europe. Vectors 10(1):357. https://doi.org/10.1186/s13071-017-2292-4
Sleeckx N, Van Gorp S, Koopman R, Kempen I, Van Hoye K, De Baere K, Zoons J, De Herdt P (2019) Production losses in laying hens during infestation with the poultry red mite Dermanyssus gallinae. Avian Pathol 48(1):17–21. https://doi.org/10.1080/03079457.2019.1641179
Sokołowicz Z, Krawczyk J, Dykiel M (2018) The effect of the type of alternative housing system, genotype and age of laying hens on egg quality. Ann Anim Sci 18(2):541–555. https://doi.org/10.2478/aoas-2018-0004
Sparagano OAE, George DR, Harrington DWJ, Giangaspero A (2014) Significance and control of the poultry red mite, Dermanyssus gallinae. Annu Rev Entomol 59:447–466. https://doi.org/10.1146/annurev-ento-011613-162101
Stackebrandt E, Koch C, Gvozdiak O, Schumann P (1995) Taxonomic dissection of the genus Micrococcus: Kocuria gen. nov., Nesterenkonia gen. nov., Kytococcus gen. nov., Dermacoccus gen. nov., and Micrococcus Cohn 1872 gen. emend. Int J Syst Evol 45(4):682–692. https://doi.org/10.1099/00207713-45-4-682
Vallalar B 2012 Investigation of the growth and survival of bacteria from Mars analog environments when exposed to Mars-like conditions. https://digitalcommons.lsu.edu/cgi/viewcontent.cgi?article=4600&context=gradschool_theses. Accessed 30 July 2022.
Valiente Moro C, De Luna CJ, Tod A, Guy JH, Sparagano OAE, Zenner L (2009) The poultry red mite (Dermanyssus gallinae): a potential vector of pathogenic agents. Exp Appl Acarol 48:93–104. https://doi.org/10.1007/s10493-009-9248-0
Valiente Moro C, Thioulouse J, Chauve C, Zenner L (2011) Diversity, geographic distribution, and habitat-specific variations of microbiota in natural populations of the chicken mite, Dermanyssus gallinae. J Med Entomol 48(4):788–796. https://doi.org/10.1603/ME10113
Weinert LA, Araujo-Jnr EV, Ahmed MZ, Welch JJ (2015) The incidence of bacterial endosymbionts in terrestrial arthropods. Proc R Soc B 282:20150249. https://doi.org/10.1098/rspb.2015.0249
Weisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703. https://doi.org/10.1128/jb.173.2.697-703.1991
Weiss B, Aksoy S (2011) Microbiome influences on insect host vector competence. Trends Parasitol 27(11):514–522. https://doi.org/10.1016/j.pt.2011.05.001
Werren J, Baldo L, Clark M (2008) Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol 6:741–751. https://doi.org/10.1038/nrmicro1969
Zabalou S, Riegler M, Theodorakopoulou M, Stauffer C, Savakis C, Bourtzis K (2004) Wolbachia-induced cytoplasmic incompatibility as a means for insect pest population control. PNAS 101(42):15042–15045. https://doi.org/10.1073/pnas.0403853101
Zchori-Fein E, Perlman SJ (2004) Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol 13:2009–2201. https://doi.org/10.1111/j.1365-294X.2004.02203.x
Zindel R, Gottlieb Y, Aebi A (2011) Arthropod symbioses: a neglected parameter in pest-and disease-control programmes. J Appl Ecol 48(4):864–872. https://doi.org/10.1111/j.1365-2664.2011.01984.x
Acknowledgments
We would like to thank Dr. Jan Hubert for his helpful advice on the interpretation and analysis of the results.
Funding
A part of this work was supported by the Ankara University Scientific Research Projects Coordination Unit (grant number 21H0239002).
Author information
Authors and Affiliations
Contributions
Nafiye Koç and Serpil Nalbantoğlu conceived the ideas. Nafiye Koç collected the samples and performed the experiments. All the authors analyze the data. Nafiye Koç wrote the main manuscript text as well as prepared the tables and figures. Both the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
The authors declare that they have participated in this work.
Consent for publication
The authors declare that they know the content of this manuscript and agree to submit it to Parasitology Research.
Competing interest
The authors declare no competing interests.
Additional information
Section Editor: Van Lun Low
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information

Fig. S1
Map of research area showing sampling sites of 21 Dermanyssus gallinae populations. The stars identify the samples that used microbiome analyses and the colors, orange; green; and blue indicate C1, F1; C2, F2; and C3, F3, respectively. (PNG 821 kb)

Fig. S2
Haplotype network analysis based on COI sequences of worldwide samples of Dermanyssus gallinae. The haplotypes obtained in the present study have been indicated by the arrow. (PNG 110 kb)

Fig. S3
Principal Component Analysis; red marks show cages, and blue marks indicate free-range farms. (PNG 149 kb)
Table S1
(DOCX 16 kb)
Table S2
(DOCX 17.8 kb)
Table S3
(XLSX 28 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Koç, N., Nalbantoğlu, S. Microbiome comparison of Dermanyssus gallinae populations from different farm rearing systems and the presence of common endosymbiotic bacteria at developmental stages. Parasitol Res 122, 227–235 (2023). https://doi.org/10.1007/s00436-022-07721-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-022-07721-2