Abstract
Cryptosporidium spp. are worldwide protozoan parasites that can affect to a broad range of vertebrate hosts, including rodents. In the island of Corsica (France), there are no previous data about these protozoa infecting wild rodents. To estimate the distribution and occurrence, a total of 117 wild murine rodents of the species Rattus rattus (84), Mus musculus domesticus (21), Apodemus sylvaticus (11), and Rattus norvegicus (1) were captured in 24 different biotopes. Fecal samples were screened for Cryptosporidium spp. by nested PCR to amplify an 830 bp fragment of the 18S rRNA gene. As general occurrence, 15.4% of the rodents analyzed were positive for Cryptosporidium spp., being detected widely distributed along the island in R. rattus (17.6%) and M. m. domesticus (14.3%). Cryptosporidium viatorum, Cryptosporidium sp. rat genotype II, and Cryptosporidium sp. rat genotype III were successfully identified in R. rattus. The results herein reported provide the first data on Cryptosporidium spp. in wild murine species from a Mediterranean island and constitute the first report of the zoonotic species C. viatorum in R. rattus. Although a low occurrence of Cryptosporidium spp. in murids was obtained and only in one animal the zoonotic species C. viatorum was identified, our results highlight that wild murine rodents from Corsica could mediate in the maintenance and transmission of this protozoan to the environment and other hosts including humans and animals. Further studies are required to better understand the epidemiology of Cryptosporidium spp. in wild rodents from Corsica and their possible public health repercussions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The protozoan Cryptosporidium spp. (phylum Apicomplexa) infects a broad range of vertebrate hosts that can play an important role in the maintenance and transmission of this pathogen (Kváč et al. 2014). It can be found in more than 150 mammalian species (Bauerfeind et al. 2016), being the order Rodentia, which represents at least 43% of the mammalian species (Wilson and Reeder 1993; Huchon et al. 2002), the most abundant and diversified order of mammals considered to be reservoirs of Cryptosporidium spp. (Feng 2010). The morphological and biological adaptations of rodents allow them to survive in any type of environments (Huchon et al. 2002), being significantly more abundant in anthropogenically modified habitats (e.g., agricultural lands, pasturelands, urban areas) than non-modified habitats (Mendoza et al. 2020). This ability facilitates rodents to spread and transfer their pathogens, such as Cryptosporidium spp., to humans and wild and domestic animals in rural and urban areas (Meerburg et al. 2009). Cryptosporidium spp. are the causal agent of cryptosporidiosis, a leading cause of diarrheal disease in both humans and animals worldwide (Innes et al. 2020). This protozoan can be transmitted by fecal–oral contamination, via ingestion of contaminated water or food, direct contact with infected persons or animals, indirect contact with contaminated fomites, and, in some cases, via inhalation (Sponseller et al. 2014; Bauerfeind et al. 2016).
The prevalence of Cryptosporidium spp. infection in rodents is highly variable worldwide (Feng 2010). A recent systematic review and meta-analysis estimated a 17% of pooled global prevalence of Cryptosporidium spp. infection in rodents, obtaining a 22% of prevalence in Europe (see Taghipour et al. 2020). In fact, previous studies reported prevalence rates of Cryptosporidium spp. infection in rodents ranging between 8.0 and 31.4% in mice and 2.1 and 63.0% in rats (Feng 2010; Koehler et al. 2018; Zhao et al. 2018, 2019). Currently, more than 40 Cryptosporidium species and similar number of genotypes have been recognized as valid worldwide (Zahedi and Ryan 2020; Ježková et al. 2020; Zahedi et al. 2021). At least 21 Cryptosporidium species and 21 genotypes have been cited in rodents: Cryptosporidium muris, Cryptosporidium parvum, Cryptosporidium hominis, Cryptosporidium meleagridis, Cryptosporidium tyzzeri, Cryptosporidium ubiquitum, Cryptosporidium suis, Cryptosporidium scrofarum, Cryptosporidium erinacei, Cryptosporidium canis, Cryptosporidium wrairi, Cryptosporidium rubeyi, Cryptosporidium andersoni, Cryptosporidium proliferans, Cryptosporidium occultus, Cryptosporidium viatorum, Cryptosporidium ditrichi, Cryptosporidium apodemi, Cryptosporidium alticolis, Cryptosporidium microti, Cryptosporidium ratti, rat genotypes II–IV, mouse genotypes II and III, naruko genotype, ferret genotype, chipmunk genotypes I and II, skunk genotype, hamster genotype, deer mouse genotypes I–IV, vole genotype, bear genotype, muskrat genotypes I and II, and ground squirrel genotypes I–III (see Lv et al. 2009; Paparini et al. 2012; Ng-Hublin et al. 2013; Kváč et al. 2014; Čondlová et al. 2018; Koehler et al. 2018; Zhang et al. 2018; Zhao et al. 2018; Horčičková et al. 2019; Zahedi and Ryan 2020; Ježková et al. 2020). Among them, C. parvum and C. hominis are the major species involved in human and waterborne outbreaks (Xiao 2010; Ryan and Xiao 2014). Cryptosporidium parvum, C. muris, and Cryptosporidium sp. rat genotype III are the species frequently detected in rats (Koehler et al. 2018), being C. muris and Cryptosporidium sp. mouse genotype I the most reported species in mice (Morgan et al. 1999, 2000; Foo et al. 2007).
Corsica (France), considered the fourth largest Mediterranean island and the most mountainous and forested one (Grech-Angelini et al. 2016), is situated in the southeast of the French mainland and west of the Italian Peninsula (Fig. 1). This well-known tourist destination is characterized by a mild Mediterranean climate and a high variability of microclimates (Grech-Angelini et al. 2016), which allows the development of a large diversity of natural environments where many animal species, including rodents, are well established. In terms of public health and veterinary, available data about the pathogens harboring wild murine rodents from Corsica are based on Fasciola hepatica in Rattus rattus and Mus musculus (Valero et al. 1998, 2002; Ménard et al. 2000; Magnanou et al. 2006), Schistosoma spp. in R. rattus (Oleaga et al. 2019), and Borrelia burgdorferi sl in Ixodes ricinus ticks of R. rattus (Cicculli et al. 2019). Within a multidisciplinary study, the species Coxiella burnetii and Toxoplasma gondii in R. rattus and M. m. domesticus (Izquierdo-Rodríguez et al. 2019) and pathogenic Leptospira species in R. rattus, Mus musculus domesticus, Rattus norvegicus, and Apodemus sylvaticus (Izquierdo-Rodríguez et al. 2020) were detected. As a part of this multidisciplinary study, and considering the lack of data on Cryptosporidium spp. in rodents from Corsica, we aimed to analyze its distribution, occurrence, and identity of the species and genotypes present in wild murids from Corsica.
Geographical location of Corsica (France) and map of the island showing the distribution of rodent sampling locations (numbers 1–24). The presence of Cryptosporidium spp. in wild rodents are represented in a black circle. (The original images were taken from d-maps (https://d-maps.com/carte.php?num_car=2801&lang=es, https://d-maps.com/carte.php?num_car=2233&lang=es) in which the original author authorized its free use for any purpose. Both images were edited by Paint 3D program)
Materials and methods
Sample collection
From a multidisciplinary study carried out in Corsica (France) between February and June 2016, a total of 117 wild murine rodents of the species R. rattus (black rat) (n = 84), M. m. domesticus (domestic mouse) (n = 21), A. sylvaticus (wood mouse) (n = 11), and R. norvegicus (gray rat) (n = 1) were captured alive using live traps. With the aim of collecting animals from different biotopes, traps were set in 24 municipalities along Corsica considering North and South of the island following Izquierdo-Rodríguez and collaborators (Izquierdo-Rodríguez et al. 2020) (Table 1), mainly at suburban-rural sites near ponds, river mouths, and lakes (Fig. 1). Euthanasia was performed with CO2 inhalation or by cervical dislocation, and, after dissection, a portion of fecal samples from the rectum of each animal were preserved in 2.5% potassium dichromate and conserved until examination. This study was carried out in strict accordance with the recommendations of the guidelines of animal welfare in experimental science and the European Union legislation (Directive 86/609/EEC). The protocol was approved by the Ethics Committee of Investigation and Animal Wellness of Universidad de La Laguna (register number CEIBA2018-0330).
DNA isolation
Before DNA extraction, 200 µl of the samples was washed with PBS-EDTA at room temperature to remove the potassium dichromate. Next, total DNA was isolated directly by following the manufacturer’s instructions of the commercial FastDNA SPIN kit (Qbiogene, Illkirch Cedex, France) using the TissueLyser II (Qiagen, Hilden, Germany) as oocyst disruptor. DNA was stored at − 20 °C until further processing.
PCR amplification
Cryptosporidium spp. were detected by nested PCR targeting an 830 bp of the 18S ribosomal rRNA gene using the primer pair SSU-F1/SSU-R1 and SSU-F2/SSU-R2 for the primary and secondary PCR, respectively (Zhao et al. 2013). The set-up of the PCR reactions was carried out according to the method of Zhang and collaborators (Zhang et al. 2015). The reaction mixture for all pairs of primers contained 0.625 U Taq DNA polymerase, 0.4 µM of each primer, 200 µM each dNTPs, 2 mM MgCl2, 1 × buffer (Mg2 + free), 2 µl of DNA template, and water to a total volume of 25 µl. The cycling conditions for both amplifications were initial denaturation of 95 °C for 5 min followed by 35 cycles 94 °C for 45 s, 45 s at suitable temperature (55 °C for primary PCR and 58 °C for secondary PCR), and 1 min at 72 °C, followed by a final extension step at 72 °C for 10 min. All PCR reactions were performed in a Labnet Thermocycler (Labnet International, Berkshire, UK). PCR products were resolved on 1.5% agarose gels, and PCR-positive products were purified using the UltraClean PCR Clean-up kit (Mo-Bio Laboratories, Inc., Carlsbad, CA, USA). In cases where bands of different sizes occurred, the desired size band was cut and purified with the QIAEX® II Gel Extraction kit (Qiagen, Hilden, Germany). In both cases, the manufacturer’s recommendations were followed.
Sequencing and phylogenetic analyses
The purified PCR-positive products were sequenced at Macrogen Europe (Amsterdam, the Netherlands). Nucleotide sequences obtained were edited with the MEGA X program (Kumar et al. 2018) and subsequently aligned with the ClustalW program included in MEGA X. Minor corrections, to increase the aligned sequence similarity and improve the inferences on any positional homology, were then made by hand. A BLAST search was carried out in order to elucidate any homologies or similarities with the sequences previously published in the GenBank database. The molecular identification was achieved by phylogenetic analysis through the neighbor-joining distance method (Saitou and Nei 1987) with at least 1000 bootstrap replications. Nucleotide sequences obtained in this work and reference sequences from other Cryptosporidium species and genotypes from the GenBank were aligned. Toxoplasma gondii was used as the outgroup. The obtained sequences generated in this study have been deposited in GenBank under the accession numbers MW590662–MW590669.
Statistical analyses
A chi-square test, setting the P value in 0.05, was conducted for the comparison of Cryptosporidium spp. prevalence obtained between the rodent species and the capture area, North and South. Data analysis was carried out using SPSS v22.0 statistical software.
Results
In this study, Cryptosporidium DNA was detected in wild murids fecal samples, and it was found widely distributed along Corsica, being detected both in the North and the South of the island homogeneously with 11.2% and 28.6% of occurrence, respectively, without statistical differences (Fig. 1, Table 1). Of the 117 wild murine rodents analyzed, 18 resulted positive for the amplification of the18S rRNA gene fragment with a general occurrence of 15.4% (Table 1). Considering the four murine species analyzed, R. rattus and M. m. domesticus were the only species found infected by Cryptosporidium spp. Of the 18 PCR-positive samples, 15 correspond to rats, nine of them were captured in the North of Corsica, specifically in the regions of Diane (n = 2), Gradugine (n = 1), Foce (n = 2), Corte (n = 2), and Biguglia (n = 2). The other six rats were captured in the South regions of Porticcio (n = 2), Ajaccio (n = 1), Pinarellu (n = 1), and Rizzanese (n = 2). In the case of mice, three resulted infected by Cryptosporidium spp., two of them were captured in the South region of Pinarellu and the other one was captured in the North region of Golo (Fig. 1). No significant differences were found in the occurrence of murine species infected between North and South of Corsica. The general occurrences of infection in R. rattus and M. m. domesticus were 17.6% (15/85) and 14.3% (3/21), respectively (Table 1).
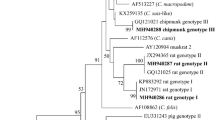
In this study, only eight PCR-positive samples from R. rattus were successfully sequenced (Table 1). Phylogenetic analysis showed the presence of 3 Cryptosporidium species and genotypes (Fig. 2), including Cryptosporidium sp. rat genotype II, Cryptosporidium sp. rat genotype III, and the zoonotic species C. viatorum. Cryptosporidium sp. rat genotype II was the most detected genotype, which has been found widely distributed in North and South of Corsica, while Cryptosporidium sp. rat genotype III was only detected in one rat in the South, as was C. viatorum. A sequence obtained from R. rattus, captured in the South region of Rizzanese (isolate 111C), clustered together with a C. viatorum sequence obtained from Berylmys bowersi from China (GenBank: MK522270). Additionally, six sequences from R. rattus, captured in the North regions of Diane (isolates 7C and 10C), Corte (isolate 89C), and Biguglia (isolate 105C), and in the South regions of Ajaccio (isolate 77C) and Pinarellu (isolate 98C), clustered together with Cryptosporidium sp. rat genotype II from Rattus tanezumi from China (GenBank: GQ121025). One sequence from R. rattus, captured in the South region of Rizzanese (isolate 113C), clustered together with Cryptosporidium sp. rat genotype III from R. rattus from Australia (GenBank: JX294371).
Four different haplotypes of Cryptosporidium sp. rat genotype II were detected. The most prevalent, haplotype 1, was found in the North (isolates 10C and 89C) and South (isolate 77C) of Corsica. One second haplotype, haplotype 2, was identified in the North of the island (isolate 105C), whose nucleotide sequence presented two extra adenine bases in positions 172 and 173 with respect to haplotype 1. In the case of haplotype 3, detected in Southern Corsica (isolate 98C), it presented a base pair change of T→A and C→A at position 63 and 235 with respect to haplotype 1, respectively. In the haplotype 4, detected in Northern Corsica (isolate 7C), the nucleotide sequenced presented a base pair change of T→G in position 160, A→C in position 161, and C→G in positions 158 and 246.
Discussion
This is the first epidemiology survey of Cryptosporidium spp. in murine rodents from Corsica. A total of three Cryptosporidium species and genotypes, concretely, Cryptosporidium sp. rat genotype II, Cryptosporidium sp. rat genotype III, and C. viatorum, were successfully identified with a 15.4% of occurrence. The occurrences obtained for rats (17.6%) and mice (14.3%) are within the reported ranges for mice (8.0–31.4%) and rats (2.1–63.0%) (Feng 2010; Koehler et al. 2018; Zhao et al. 2018, 2019).
The most prevalent genotype was Cryptosporidium sp. rat genotype II, contrary to the genotype found in other studies where Cryptosporidium sp. rat genotype III is the most frequently reported (Koehler et al. 2018). Cryptosporidium sp. rat genotype II was found in six rats widely distributed both in the North and South of the island of Corsica. However, Cryptosporidium sp. rat genotype III and C. viatorum were only found in the South of the island, but as they have been detected in a single animal each, this is not a congruent result to assess if there are any difference between their distribution on the island. Therefore, more studies are required to search for these species and genotypes in the North of the island, considering the few animals from which we were able to successfully sequence Cryptosporidium spp.
Cryptosporidium sp. rat genotype II and Cryptosporidium sp. rat genotype III are host-adapted genotypes (Zahedi et al. 2016) that have been detected in rats worldwide (Lv et al. 2009; Paparini et al. 2012; Ng-Hublin et al. 2013; Koehler et al. 2018; Tan et al. 2019; Zhao et al. 2019; García-Livia et al. 2020). Nevertheless, other hosts can be infected by Cryptosporidium sp. rat genotypes II and III, as has been reported in mice, cats, sheep, and goats (Ryan et al. 2005; Lv et al. 2009; Paparini et al. 2012; Ng-Hublin et al. 2013; Koinari et al. 2014; Yang et al. 2015; Hatam-Nahavandi et al. 2019). To date, the potential of the rat genotypes identified in this study to cause cryptosporidiosis in livestock or humans remains unclear.
Cryptosporidium viatorum, the only zoonotic species identified in this study, has been originally found exclusively in humans (Elwin et al. 2012; Insulander et al. 2013; Lebbad et al. 2013; Adamu et al. 2014; Ayinmode et al. 2014; Stensvold et al. 2015; De Lucio et al. 2016; Sánchez et al. 2017; Ukwah et al. 2017) and in urban wastewater and combined sewer overflows in China (Huang et al. 2017). Recently in 2018, C. viatorum was reported in a non-human host, concretely in the wild rats Rattus lutreolus from Australia (Koehler et al. 2018), and in the species R. norvegicus, Leopoldamys edwardsi, and B. bowersi from China (Zhao et al. 2019; Chen et al. 2019). Therefore, our study constitutes the first report of C. viatorum in R. rattus and the fifth report in non-human hosts, expanding its host range. Our results support recent findings suggesting that wild murines could be hosts of the zoonotic species C. viatorum. In fact, C. viatorum XVa subtype family from wild rats has been recently reported to be genetically identical to those subtypes found in humans (Chen et al. 2019). Further molecular investigations are needed to better understand the epidemiology of C. viatorum and clarify its transmission routes.
Considering that the species C. viatorum was the only zoonotic species detected in a single animal, more analyses are required to determine the occurrence of this and other possible zoonotic Cryptosporidium species that could have not been detected in wild murids from Corsica. In this sense, it is important to note that a recent study, conducted between 2017 and 2019 with 750 online reports from the National Reference Center-Expert of cryptosporidiosis in France, showed that the 40% of the reported cases of human cryptosporidiosis in this country were mainly related to recreational water (48%) and animal contact (23%) (Costa et al. 2020). In Corsica, no cases of cryptosporidiosis in humans have been reported to date, but considering that murine rodents can maintain the transmission cycles of several Cryptosporidium zoonotic species, such as C. parvum, C. muris, C. ubiquitum, C. meleagridis, C. scrofarum, C. viatorum, C. canis, C. tyzzeri, C. andersoni, C. hominis, C. suis, C. proliferans, C. occultus, C. wrairi, C. rubeyi, and C. ditrichi (Zhao et al. 2018; Koehler et al. 2018; Zhang et al. 2018; Beser et al. 2020), the detection of C. viatorum in this study should not be dismissed.
This study constitutes the first survey of Cryptosporidium spp. infection in wild murine species from a Mediterranean island. Some studies have previously confirmed the presence of this protozoan in murine rodent species from Atlantic islands (Feliu et al. 2012; García-Livia et al. 2020), showing that Cryptosporidium spp. infection is common in R. rattus and M. m. domesticus from the Canary Islands (Spain), with a high diversity of Cryptosporidium species and genotypes. In Tenerife (Canary Islands), a total of seven Cryptosporidium species/genotypes were identified, while in Corsica, despite being an island much larger in size than Tenerife and both studies with similar sample size (n = 97 in Tenerife and n = 1117 in Corsica), a low occurrence was obtained and a total of three Cryptosporidium species/genotypes were identified. Further studies analyzing more samples are required in order to obtain additional data about the occurrence and biodiversity of Cryptosporidium spp. in wild murids from Corsica.
Factors such as host specificity, ecological interactions, geographic distribution, bioclimatic conditions, and sampled areas, among others, could influence in the occurrence and prevalence of Cryptosporidium spp. in wild rodents (Zhao et al. 2019). Considering that samples of our study were only obtained near ponds, river mouths, and lakes, together with the fact that only few samples were successfully sequenced, further investigations with an increased in the number of samples and in non-analyzed areas would improve the knowledge about the prevalence, distribution, and biodiversity of Cryptosporidium spp. in wild murine rodents from Corsica.
Conclusions
The present study constitutes the first report of Cryptosporidium spp. in wild murids from Corsica (France), thus creating an overview of the epidemiological situation of this parasite in this region. Cryptosporidium sp. rat genotype II, Cryptosporidium sp. rat genotype III, and C. viatorum were the Cryptosporidium species and genotypes successfully identified in fecal samples from wild murine rodents, being the first citation of C. viatorum in R. rattus.
Considering the interactions between wildlife, livestock, and humans, further investigations should be carried out in unsampled areas, in addition to other wild hosts, to better understand the epidemiology of Cryptosporidium spp. in Corsica and to determine the possible zoonotic risks of transmission.
Availability of data and material
The dataset used and analyzed by the authors during the present study are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, Feng Y, Xiao L (2014) Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PloS Negl Trop Dis 8:e2831. https://doi.org/10.1371/journal.pntd.0002831
Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L (2014) Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health 61:297–303. https://doi.org/10.1111/zph.12072
Bauerfeind R, Von Graevenitz A, Kimmig P, Schiefer HG, Schwarz T, Slenczka W, Zahner H (2016) Parasitic zoonoses. In: Bauerfeind R, Von Graevenitz A, Kimmig P, Schiefer HG, Schwarz T, Slenczka W, Zahner H (eds) Zoonoses: infectious diseases transmissible from animals to humans, 4th edn. ASM Press, Washington DC, USA, pp. 324–327
Beser J, Bujila I, Wittesjö B, Lebbad M (2020) From mice to men: three cases of human infection with Cryptosporidium ditrichi. Infect Genet Evol 78:104120. https://doi.org/10.1016/j.meegid.2019.104120
Chen YW, Zheng WB, Zhang NZ, Gui BZ, Lv QY, Yan JQ, Zhao Q, Liu GH (2019) Identification of Cryptosporidium viatorum XVa subtype family in two wild rat species in China. Parasit Vectors 12:502. https://doi.org/10.1186/s13071-019-3763-6
Cicculli V, Capai L, Quilichini Y, Masse S, Fernández-Alvarez A, Minodier L, Bompard P, Charrel R, Falchi A (2019) Molecular investigation of tick-borne pathogens in ixodid ticks infesting domestic animals (cattle and sheep) and small rodents (black rats) of Corsica, France. Ticks Tick Borne Dis 10:606–613. https://doi.org/10.1016/j.ttbdis.2019.02.007
Čondlová Š, Horčičková M, Sak B, Květoňová D, Hlásková L, Konečný R, Stanko M, McEvoy J, Kváč M (2018) Cryptosporidium apodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur J Protistol 63:1–12. https://doi.org/10.1016/j.ejop.2017.12.006
Costa D, Razakandrainibe R, Valot S, Vannier M, Sautour M, Basmaciyan L, Gargala G, Viller V, Lemeteil D, Ballet JJ, French National Network on Surveillance of Human Cryptosporidiosis, Dalle F, Favennec L (2020) Epidemiology of cryptosporidiosis in France from 2017 to 2019. Microorganisms 8:1358. https://doi.org/10.3390/microorganisms8091358
De Lucio A, Amor-Aramendı́a A, Bailo B, Saugar JM, Anegagrie M, Arroyo A, López-Quintana B, Zewdie D, Ayehubizu Z, Yizengaw E, Abera B, Yimer M, Mulu W, Hailu T, Herrador Z, Fuentes I, Carmena D (2016) Prevalence and genetic diversity of Giardia duodenalis and Cryptosporidium spp. among school children in a rural area of the Amhara region, North-West Ethiopia. PLoS One 11:e0159992. https://doi.org/10.1371/journal.pone.0159992
Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM (2012) Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol 42:675–682. https://doi.org/10.1016/j.ijpara.2012.04.016
Feliu C, López M, Gómez MS, Torres J, Sánchez S, Miquel J, Abreu-Acosta N, Segovia JM, Martín-Alonso A, Montoliu I, Villa M, Fernández-Álvarez A, Bakhoum AJS, Valladares B, Orós J, Foronda P (2012) Parasite fauna of rodents (Murinae) from El Hierro (Canary Islands, Spain): a multidisciplinary approach. Acta Parasitol 57:171–178. https://doi.org/10.2478/s11686-012-0016-7
Feng Y (2010) Cryptosporidium in wild placental mammals. Exp Parasitol 124:128–137. https://doi.org/10.1016/j.exppara.2008.11.005
Foo C, Farrell J, Boxell A, Robertson I, Ryan UM (2007) Novel Cryptosporidium genotype in wild Australian mice (Mus domesticus). Appl Environ Microbiol 73:7693–7696. https://doi.org/10.1128/AEM.00848-07
García-Livia K, Martín-Alonso A, Foronda P (2020) Diversity of Cryptosporidium spp. in wild rodents from the Canary Islands, Spain. Parasit Vectors 13:445. https://doi.org/10.1186/s13071-020-04330-9
Grech-Angelini S, Stachurski F, Lancelot R, Boissier J, Allienne JF, Marco S, Maestrini O, Uilenberg G (2016) Ticks (Acari: Ixodidae) infesting cattle and some other domestic and wild hosts on the French Mediterranean island of Corsica. Parasit Vectors 9:582. https://doi.org/10.1186/s13071-016-1876-8
Hatam-Nahavandi K, Ahmadpour E, Carmena D, Spotin A, Bangoura B, Xiao L (2019) Cryptosporidium infections in terrestrial ungulates with focus on livestock: a systematic review and meta-analysis. Parasit Vectors 12:453. https://doi.org/10.1186/s13071-019-3704-4
Horčičková M, Čondlová Š, Holubová N, Sak B, Květoňová D, Hlásková L, Konečný R, Sedláček F, Clark M, Giddings C, McEvoy J, Kváč M (2019) Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. (Apicomplexa: Cryptosporidiidae). Parasitology 146:220–233. https://doi.org/10.1017/S0031182018001142
Huang C, Hu Y, Wang L, Wang Y, Li N, Guo Y, Feng Y, Xiao L (2017) Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. App Environ Microbiol 83:e00682-e717. https://doi.org/10.1128/AEM.00682-17
Huchon D, Madsen O, Sibbald MJJB, Ament K, Stanhope MJ, Catzeflis F, De Jong WW, Douzery EJP (2002) Rodent phylogeny and a timescale for the evolution of Glires: evidence from an extensive taxon sampling using three nuclear genes. Mol Biol Evol 19:1053–1065. https://doi.org/10.1093/oxfordjournals.molbev.a004164
Innes EA, Chalmers RM, Wells B, Pawlowic MC (2020) A one health approach to tackle cryptosporidiosis. Trends Parasitol 36:290–303. https://doi.org/10.1016/j.pt.2019.12.01
Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B (2013) Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect 141:1009–1020. https://doi.org/10.1017/S0950268812001665
Izquierdo-Rodríguez E, Fernández-Álvarez A, Martín-Carrillo N, Feliu C, Marchand B, Quilichini Y, Foronda P (2019) Rodents as reservoirs of the zoonotic pathogens Coxiella burnetii and Toxoplasma gondii in Corsica (France). Vector Borne Zoonotic Dis 19:879–883. https://doi.org/10.1089/vbz.2019.2485
Izquierdo-Rodríguez E, Fernández-Álvarez A, Martín-Carrillo N, Marchand B, Feliu C, Miquel J, Foronda P, Quilichini Y (2020) Pathogenic Leptospira species in rodents from Corsica (France). PLoS One 15:e0233776. https://doi.org/10.1371/journal.pone.0233776
Ježková J, Prediger J, Holubová N, Sak B, Konečný R, Feng Y, Xiao L, Rost M, McEvoy J, Kváč M (2020) Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology 28:1–14. https://doi.org/10.1017/S0031182020001833
Koehler AV, Wang T, Haydon SR, Gasser RB (2018) Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus - an emerging zoonotic pathogen? Int J Parasitol Parasites Wildl 7:18–26. https://doi.org/10.1016/j.ijppaw.2018.01.004
Koinari M, Lymbery AJ, Ryan UM (2014) Cryptosporidium species in sheep and goats from Papua New Guinea. Exp Parasitol 141:134–137. https://doi.org/10.1016/j.exppara.2014.03.021
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kváč M, McEvoy J, Stenger B, Clark M (2014) Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G (eds) Cryptosporidium: Parasite and Disease. Springer, Vienna, pp 237–326
Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, Axen C (2013) Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology 140:1735–1740. https://doi.org/10.1017/S003118201300084X
Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, Wang H, Feng C, Wang X, Ren X, Qi M, Xiao L (2009) Cryptosporidium spp. in wild, laboratory, and pet rodents in China: prevalence and molecular characterization. Appl Environ Microbiol 75:7692–7699. https://doi.org/10.1128/AEM.01386-09
Magnanou E, Fons R, Feliu C, Morand S (2006) Physiological responses of insular wild black rat (Rattus rattus) to natural infection by the digenean trematode Fasciola hepatica. Parasitol Res 99:97–101. https://doi.org/10.1007/s00436-005-0063-1
Meerburg BG, Singleton GR, Kijlstra A (2009) Rodent-borne diseases and their risks for public health. Crit Rev Microbiol 35:221–270. https://doi.org/10.1080/10408410902989837
Ménard A, l’Hostis M, Leray G, Marchandeau S, Pascal M, Roudot N, Michel V, Chauvin A (2000) Inventory of wild rodents and lagomorphs as natural hosts of Fasciola hepatica on a farm located in a humid area in Loire Atlantique (France). Parasite 7:77–82. https://doi.org/10.1051/parasite/2000072077
Mendoza H, Rubio AV, García-Peña GE, Suzán G, Simonetti JA (2020) Does land-use change increase the abundance of zoonotic reservoirs? Rodents say yes. Eur J Wildl Res 66:6. https://doi.org/10.1007/s10344-019-1344-9
Morgan UM, Sturdee AP, Singleton G, Gomez MS, Gracenea M, Torres J, Hamilton SG, Woodside DP, Thompson RCA (1999) The Cryptosporidium “Mouse” genotype is conserved across geographic areas. J Clin Microbiol 37:1302–1305. https://doi.org/10.1128/JCM.37.5.1302-1305.1999
Morgan UM, Xiao L, Monis P, Sulaiman I, Pavlasek I, Blagburn B, Olson M, Upton SJ, Khramtsov NV, Lal A, Elliot A, Thompson RCA (2000) Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology 120:457–464. https://doi.org/10.1017/S0031182099005703
Ng-Hublin JS, Singleton GR, Ryan U (2013) Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect Genet Evol 16:5–12. https://doi.org/10.1016/j.meegid.2013.01.011
Oleaga A, Rey O, Polack B, Grech-Angelini S, Quilichini Y, Pérez-Sánchez R, Boireau P, Mulero S, Brunet A, Rogon A, Vallée I, Kincaid-Smith J, Allienne JF, Boissier J (2019) Epidemiological surveillance of schistosomiasis outbreak in Corsica (France): are animal reservoir hosts implicated in local transmission? PLoS Negl Trop Dis 13:e0007543. https://doi.org/10.1371/journal.pntd.0007543
Paparini A, Jackson B, Ward S, Young S, Ryan UM (2012) Multiple Cryptosporidium genotypes detected in wild black rats (Rattus rattus) from northern Australia. Exp Parasitol 131:404–412. https://doi.org/10.1016/j.exppara.2012.05.009
Ryan U, Xiao L (2014) Taxonomy and molecular taxonomy. In: Cacciò SM, Widmer G (eds) Cryptosporidium: parasite and disease. Springer, Vienna, pp 3–41
Ryan UM, Bath C, Robertson I, Read C, Elliot A, Mcinnes L, Traub R, Besier B (2005) Sheep may not be an important zoonotic reservoir for Cryptosporidium and Giardia parasites. Appl Environ Microbiol 71:4992–4997. https://doi.org/10.1128/AEM.71.9.4992-4997.2005
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sánchez A, Munoz M, Gómez N, Tabares J, Segura L, Salazar A, Restrepo C, Ruíz M, Reyes P, Qian Y, Xiao L, Lopez MC, Ramirez JD (2017) Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among indigenous children from the Colombian amazon basin. Front Microbiol 8:248. https://doi.org/10.3389/fmicb.2017.00248
Sponseller JK, Griffiths JK, Tzipori S (2014) The evolution of respiratory cryptosporidiosis: evidence for transmission by inhalation. Clin Microbiol Rev 27:575–586. https://doi.org/10.1128/CMR.00115-13
Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M (2015) Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol 53:1891–1897. https://doi.org/10.1128/JCM.00313-15
Taghipour A, Olfatifar M, Foroutan M, Bahadory S, Malih N, Norouzi M (2020) Global prevalence of Cryptosporidium infection in rodents: a systematic review and meta-analysis. Prev Vet Med 182:105119. https://doi.org/10.1016/j.prevetmed.2020.105119
Tan TK, Low VL, Ng WH, Ibrahim J, Wang D, Tan CH, Chellappan S, Lim YAL (2019) Occurrence of zoonotic Cryptosporidium and Giardia duodenalis species/genotypes in urban rodents. Parasitol Int 69:110–113. https://doi.org/10.1016/j.parint.2018.12.007
Ukwah BN, Ezeonu IM, Ezeonu CT, Roellig D, Xiao L (2017) Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J Infect Dev Ctries 11:173–179. https://doi.org/10.3855/jidc.8034
Valero MA, Marcos MD, Fons R, Mas-Coma S (1998) Fasciola hepatica development in the experimentally infected black rat Rattus rattus. Parasitol Res 84:188–194. https://doi.org/10.1007/s004360050381
Valero MA, Panova M, Comes AM, Fons R, Mas-Coma S (2002) Patterns in size and shedding of Fasciola hepatica eggs by naturally and experimentally infected murid rodents. J Parasitol 88:308–313. https://doi.org/10.1645/0022-3395(2002)088[0308:PISASO]2.0.CO;2
Wilson DE, Reeder DM (1993) Mammal species of the world: a taxonomic and geographic reference. The Johns Hopkins University Press, Baltimore
Xiao L (2010) Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol 124:80–89. https://doi.org/10.1016/j.exppara.2009.03.018
Yang R, Ying JLJ, Monis P, Ryan U (2015) Molecular characterisation of Cryptosporidium and Giardia in cats (Felis catus) in Western Australia. Exp Parasitol 155:13–18. https://doi.org/10.1016/j.exppara.2015.05.001
Zahedi A, Ryan U (2020) Cryptosporidium–an update with an emphasis on foodborne and waterborne transmission. Res Vet Sci 132:500–512. https://doi.org/10.1016/j.rvsc.2020.08.002
Zahedi A, Paparini A, Jian F, Robertson I, Ryan U (2016) Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int J Parasitol Parasites Wildl 5:88–109. https://doi.org/10.1016/j.ijppaw.2015.12.001
Zahedi A, Bolland SJ, Oskam CL, Ryan U (2021) Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exp Parasitol 223:108089. https://doi.org/10.1016/j.exppara.2021.108089
Zhang XX, Tan QD, Zhou DH, Ni XT, Liu GX, Yang YC, Zhu XQ (2015) Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol Res 114:2781–2787. https://doi.org/10.1007/s00436-015-4537-5
Zhang X, Jian Y, Li X, Ma L, Karanis G, Karanis P (2018) The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan Plateau area, China. Parasitol Res 117:1401–1407. https://doi.org/10.1007/s00436-018-5827-5
Zhao GH, Ren WX, Gao M, Bian QQ, Hu B, Cong MM, Lin Q, Wang RJ, Qi M, Qi MZ, Zhu XQ, Zhang LX (2013) Genotyping Cryptosporidium andersoni in cattle in Shaanxi Province, Northwestern China. PloS One 8:e60112. https://doi.org/10.1371/journal.pone.0060112
Zhao W, Wang J, Ren G, Yang Z, Yang F, Zhang W, Xu Y, Liu A, Ling H (2018) Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasit Vectors 11:313. https://doi.org/10.1186/s13071-018-2892-7
Zhao W, Zhou H, Huang Y, Xu L, Rao L, Wang S, Wang W, Yi Y, Zhou X, Wu Y, Ma T, Wang G, Hu X, Peng R, Yin F, Lu G (2019) Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int J Parasitol Parasites Wildl 9:317–321. https://doi.org/10.1016/j.ijppaw.2019.03.017
Acknowledgements
CF and JM are members of the 2017-SGR-1008 research group.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by post-doctoral fellowship (AFA) CE/03/2015 “Collectivité Territoriale de Corse-Direction de L’Enseignement Supérieur et de la Recherche”; ProID2017010092 Canary Council of Economy, Industry, Trade, and Knowledge and FEDER 2014–2020; and RD16/0027/0001 Spanish Ministry of Health, Consumer Affairs and Social Welfare of Spain and CEI program of Universidad de La Laguna and the Canary Council of Economy, Knowledge and Employment. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. KGL is granted a scholarship by Spanish Ministry of Science, Innovation and Universities and Universidad de La Laguna (Becas M-ULL, convocatoria 2019).
Author information
Authors and Affiliations
Contributions
KGL wrote the manuscript, designed the figures, and performed the phylogenetic analysis and the interpretation of the results. AFA has performed the experiments, formal analysis, and data curation and provided critical revision of the manuscript. CF and JM has contributed to the sampling of the rodents and provided critical revision of the manuscript. YQ has contributed with the funding acquisition and project administration and provided critical revision of the manuscript. PF has contributed with the conceptualization, funding acquisition, project administration, and interpretation of the results and drafted or provided critical revision of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was carried out in strict accordance with the recommendations of the guidelines of animal welfare in experimental science and the European Union legislation (Directive 86/609/EEC). The protocol was approved by “Comité de Ética de la Invetigación y Bienestar Animal” of Universidad de La Laguna (Protocol Number: CEIBA2018-0330).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Section Editor: Lihua Xiao
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
García-Livia, K., Fernández-Álvarez, Á., Feliu, C. et al. Cryptosporidium spp. in wild murids (Rodentia) from Corsica, France. Parasitol Res 121, 345–354 (2022). https://doi.org/10.1007/s00436-021-07369-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-021-07369-4