Abstract
Purpose
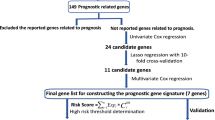
This study aims to establish a risk prediction model based on prognosis-related genes (PRGs) and clinicopathological factors, and investigate the biological activities of PRGs in lung adenocarcinoma (LUAD).
Methods
Risk score signatures were developed by employing multiple algorithms and their amalgamations. A predictive model for overall survival was established through the integration of risk score signatures and several clinicopathological parameters. A comprehensive single-cell atlas, gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) were used to investigate the biological activities of prognosis-related genes in LUAD.
Results
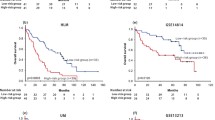
A risk prediction model was established based on 16 PRGs, exhibiting robust performance in predicting overall survival. The single-cell analysis revealed that epithelial cells were primarily associated with worse survival of LUAD, and PRGs were predominantly enriched in malignant epithelial cells and influenced epithelial cell growth and progression. Furthermore, GSEA and GSVA analysis showed that PRGs were involved in tumor pathways such as epithelial-mesenchymal transition, hypoxia and KRAS_UP, and high GSVA scores are correlated with worse outcome in LUAD patients.
Conclusions
The constructed risk prediction model in this study offers clinicians a valuable tool for tailoring treatment strategies of LUAD and provides a comprehensive interpretation on the biological activities of PRGs in LUAD.











Similar content being viewed by others
Data availability
The datasets utilized in this study are available in online repositories. The specific repository/repositories and corresponding accession number(s) can be found in the article or Supplementary Material.
Abbreviations
- AUC:
-
Area under the curve
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- GSVA:
-
Gene set variation analysis
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- OS:
-
Overall survival
- scRNA-seq:
-
Single-cell RNA sequencing
- TME:
-
Tumor microenvironment
- LUAD:
-
Lung adenocarcinoma
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene expression omnibus
- UMAP:
-
Uniform manifold approximation and projection
- PRGs:
-
Prognosis-related genes
- GBM:
-
Generalized boosted regression modeling
- EMT:
-
Epithelial–mesenchymal transition
- DCA:
-
Decision curve analysis
References
Akay MF (2009) Support vector machines combined with feature selection for breast cancer diagnosis. Expert Syst Appl 36:3240–3247. https://doi.org/10.1016/j.eswa.2008.01.009
Aran D, Looney AP, Liu L et al (2019) Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20:163–172. https://doi.org/10.1038/s41590-018-0276-y
Bilokapic S, Schwartz TU (2012) Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci 109:15241–15246. https://doi.org/10.1073/pnas.1205151109
Buscail L, Bournet B, Cordelier P (2020) Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol 17:153–168. https://doi.org/10.1038/s41575-019-0245-4
Chen J, Yang H, Teo ASM et al (2020) Genomic landscape of lung adenocarcinoma in East Asians. Nat Genet 52:177–186. https://doi.org/10.1038/s41588-019-0569-6
Choy EH, De Benedetti F, Takeuchi T et al (2020) Translating IL-6 biology into effective treatments. Nat Rev Rheumatol 16:335–345. https://doi.org/10.1038/s41584-020-0419-z
Corrêa ZM, Augsburger JJ (2016) Independent prognostic significance of gene expression profile class and largest basal diameter of posterior uveal melanomas. Am J Ophthalmol 162:20-27.e1. https://doi.org/10.1016/j.ajo.2015.11.019
Craene BD, Berx G (2013) Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer 13:97–110. https://doi.org/10.1038/nrc3447
Cruz JA, Wishart DS (2007) Applications of machine learning in cancer prediction and prognosis. Cancer Inform 2:59–77
Dai M, Pei X, Wang X-J (2022) Accurate and fast cell marker gene identification with COSG. Brief Bioinform 23:bbab579. https://doi.org/10.1093/bib/bbab579
Denisenko TV, Budkevich IN, Zhivotovsky B (2018) Cell death-based treatment of lung adenocarcinoma. Cell Death Dis 9:117. https://doi.org/10.1038/s41419-017-0063-y
Drosten M, Barbacid M (2020) Targeting the MAPK pathway in KRAS-driven tumors. Cancer Cell 37:543–550. https://doi.org/10.1016/j.ccell.2020.03.013
Eltzschig HK, Carmeliet P (2011) Hypoxia and inflammation. N Engl J Med 364:656–665. https://doi.org/10.1056/NEJMra0910283
Frazzi R (2021) BIRC3 and BIRC5: multi-faceted inhibitors in cancer. Cell Biosci 11:8. https://doi.org/10.1186/s13578-020-00521-0
Gao R, Bai S, Henderson YC et al (2021) Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol 39:599–608. https://doi.org/10.1038/s41587-020-00795-2
Gilkes DM, Semenza GL, Wirtz D (2014) Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer 14:430–439. https://doi.org/10.1038/nrc3726
Gohil SH, Iorgulescu JB, Braun DA et al (2021) Applying high-dimensional single-cell technologies to the analysis of cancer immunotherapy. Nat Rev Clin Oncol 18:244–256. https://doi.org/10.1038/s41571-020-00449-x
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
Harris IS, Treloar AE, Inoue S et al (2015) Glutathione and thioredoxin antioxidant pathways synergize to drive cancer initiation and progression. Cancer Cell 27:211–222. https://doi.org/10.1016/j.ccell.2014.11.019
Jin S, Guerrero-Juarez CF, Zhang L et al (2021) Inference and analysis of cell-cell communication using Cell Chat. Nat Commun 12:1088. https://doi.org/10.1038/s41467-021-21246-9
Johnson DE, O’Keefe RA, Grandis JR (2018) Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 15:234–248. https://doi.org/10.1038/nrclinonc.2018.8
Kessenbrock K, Plaks V, Werb Z (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141:52–67. https://doi.org/10.1016/j.cell.2010.03.015
Kidd P (2003) Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev 8:223–246
Kim N, Kim HK, Lee K et al (2020) Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun 11:2285. https://doi.org/10.1038/s41467-020-16164-1
Komorowski M, Green A, Tatham KC et al (2022) Sepsis biomarkers and diagnostic tools with a focus on machine learning. EBioMedicine 86:104394. https://doi.org/10.1016/j.ebiom.2022.104394
Korsunsky I, Millard N, Fan J et al (2019) Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16:1289–1296. https://doi.org/10.1038/s41592-019-0619-0
Langerød A, Zhao H, Borgan Ø et al (2007) TP53 mutation status and gene expression profiles are powerful prognostic markers of breast cancer. Breast Cancer Res 9:R30. https://doi.org/10.1186/bcr1675
Lg A, At E (2013) Using three machine learning techniques for predicting breast cancer recurrence. J Health Med Inform. https://doi.org/10.4172/2157-7420.1000124
Li A, Zhang C, Gao S et al (2013) TIP30 loss enhances cytoplasmic and nuclear EGFR signaling and promotes lung adenocarcinogenesis in mice. Oncogene 32:2273–2281. https://doi.org/10.1038/onc.2012.253. (2281e.1–12)
Li F, Aljahdali I, Ling X (2019) Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res 38:368. https://doi.org/10.1186/s13046-019-1362-1
Liu X, Mitchell JM, Wozniak RW et al (2012) Structural evolution of the membrane-coating module of the nuclear pore complex. Proc Natl Acad Sci USA 109:16498–16503. https://doi.org/10.1073/pnas.1214557109
Mueller SN, Gebhardt T, Carbone FR, Heath WR (2013) Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31:137–161. https://doi.org/10.1146/annurev-immunol-032712-095954
Pastushenko I, Blanpain C (2019) EMT transition states during tumor progression and metastasis. Trends Cell Biol 29:212–226. https://doi.org/10.1016/j.tcb.2018.12.001
Qiu X, Mao Q, Tang Y et al (2017) Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14:979–982. https://doi.org/10.1038/nmeth.4402
Quail DF, Joyce JA (2013) Microenvironmental regulation of tumor progression and metastasis. Nat Med 19:1423–1437. https://doi.org/10.1038/nm.3394
Quidville V, Alsafadi S, Goubar A et al (2013) Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Can Res 73:2247–2258. https://doi.org/10.1158/0008-5472.CAN-12-2501
Satija R, Farrell JA, Gennert DG et al (2015) Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33:495
Schümperli D, Pillai RS (2004) The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci 61:2560–2570. https://doi.org/10.1007/s00018-004-4190-0
Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72:7–33. https://doi.org/10.3322/caac.21708
Stelloo E, Nout RA, Osse EM et al (2016) Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin Cancer Res 22:4215–4224. https://doi.org/10.1158/1078-0432.CCR-15-2878
Sun D, Guan X, Moran AE et al (2022) Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat Biotechnol 40:527–538. https://doi.org/10.1038/s41587-021-01091-3
Suvà ML, Tirosh I (2019) Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell 75:7–12. https://doi.org/10.1016/j.molcel.2019.05.003
The Cancer Genome Atlas Research Network (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature 511:543–550. https://doi.org/10.1038/nature13385
Tong X, Li K, Luo Z et al (2009) Decreased TIP30 expression promotes tumor metastasis in lung cancer. Am J Pathol 174:1931–1939. https://doi.org/10.2353/ajpath.2009.080846
Walker JA, McKenzie ANJ (2018) TH2 cell development and function. Nat Rev Immunol 18:121–133. https://doi.org/10.1038/nri.2017.118
Wu F, Fan J, He Y et al (2021) Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun 12:2540. https://doi.org/10.1038/s41467-021-22801-0
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Yuan X, Wu H, Han N et al (2014) Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J Hematol Oncol 7:87. https://doi.org/10.1186/s13045-014-0087-z
Yunna C, Mengru H, Lei W, Weidong C (2020) Macrophage M1/M2 polarization. Eur J Pharmacol 877:173090. https://doi.org/10.1016/j.ejphar.2020.173090
Acknowledgements
We would like to express our gratitude to the researchers who generously shared the original data of open single-cell and bulk RNA sequencing in public databases. Additionally, we extend our appreciation to the providers of open-source R packages, whose contributions were instrumental in our analysis.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81773159 and 81871203).
Author information
Authors and Affiliations
Contributions
The study was designed by YM, SJY, and LYQ. YM and SJY conducted the analysis. YM, SJY, TXL, ZXY, and TDC performed the validation in the independent cohort. YM, SJY, TXL, and ZXY contributed to the preparation of figures and tables. YM and SJY drafted the manuscript. LYQ, SJY, and YY provided revisions to the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethic approval and consent to participate
This study does not involve ethical approval.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yi, M., Shi, J., Tan, X. et al. Integration and deconvolution methodology deciphering prognosis-related signatures in lung adenocarcinoma. J Cancer Res Clin Oncol 149, 16441–16460 (2023). https://doi.org/10.1007/s00432-023-05403-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05403-9