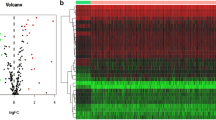
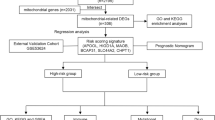
Abstract
Purpose
Mitophagy and aging (MiAg) are very important pathophysiological mechanisms contributing to tumorigenesis. MiAg-related genes have prognostic value in lung adenocarcinoma (LUAD). However, prognostic, and immune correlation studies of MiAg-related genes in LUAD are lacking.
Methods
MiAg differentially expressed genes (DEGs) in LUAD were obtained from public sequencing datasets. A prognostic model including MiAg DEGs was constructed according to patients divided into low- and high-risk groups. Gene Ontology, gene set enrichment analysis, gene set variation analysis, CIBERSORT immune infiltration analysis, and clinical characteristic correlation analyses were performed for functional annotation and correlation of MiAgs with prognosis in patients with LUAD.
Results
Seven MiAg DEGs of LUAD were identified: CAV1, DSG2, DSP, MYH11, NME1, PAICS, PLOD2, and the expression levels of these genes were significantly correlated (P < 0.05). The RiskScore of the MiAg DEG prognostic model demonstrated high predictive ability of overall survival of patients diagnosed with LUAD. Patients with high and low MiAg phenotypic scores exhibited significant differences in the infiltration levels of eight types of immune cells (P < 0.05). The multi-factor DEG regression model showed higher efficacy in predicting 5-year survival than 3- and 1-year survival of patients with LUAD.
Conclusions
Seven MiAg-related genes were identified to be significantly associated with the prognosis of patients diagnosed with LUAD. Moreover, the identified MiAg DEGs might affect the immunotherapy strategy of patients with LUAD.












Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available in the TCGA (https://portal.gdc.cancer.gov/) and GEO (GSE10072, GSE30219 and GSE3141).
References
Ali SR, Jordan M, Nagarajan P, Amit M (2022) Nerve density and neuronal biomarkers in cancer. Cancers (Basel). https://doi.org/10.3390/cancers14194817
Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R (2007) NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res 35:D760-765. https://doi.org/10.1093/nar/gkl887
Bild AH, Yao G, Chang JT et al (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439:353–357. https://doi.org/10.1038/nature04296
Bilitou A, Watson J, Gartner A, Ohnuma S (2009) The NM23 family in development. Mol Cell Biochem 329:17–33. https://doi.org/10.1007/s11010-009-0121-6
Boissan M, Wendum D, Arnaud-Dabernat S, Munier A, Debray M, Lascu I, Daniel JY, Lacombe ML (2005) Increased lung metastasis in transgenic NM23-Null/SV40 mice with hepatocellular carcinoma. J Natl Cancer Inst 97:836–845. https://doi.org/10.1093/jnci/dji143
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424. https://doi.org/10.3322/caac.21492
Cai W, van der Laan M (2020) Nonparametric bootstrap inference for the targeted highly adaptive least absolute shrinkage and selection operator (LASSO) estimator. Int J Biostat. https://doi.org/10.1515/ijb-2017-0070
Cai F, Zhu Q, Miao Y, Shen S, Su X, Shi Y (2017) Desmoglein-2 is overexpressed in non-small cell lung cancer tissues and its knockdown suppresses NSCLC growth by regulation of p27 and CDK2. J Cancer Res Clin Oncol 143:59–69. https://doi.org/10.1007/s00432-016-2250-0
Chakravarthi B, Goswami MT, Pathi SS et al (2018a) Expression and role of PAICS, a de novo purine biosynthetic gene in prostate cancer. Prostate 78:693–694. https://doi.org/10.1002/pros.23533
Chakravarthi B, Rodriguez Pena MDC, Agarwal S et al (2018b) A role for de novo purine metabolic enzyme PAICS in bladder cancer progression. Neoplasia 20:894–904. https://doi.org/10.1016/j.neo.2018.07.006
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018) Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 1711:243–259. https://doi.org/10.1007/978-1-4939-7493-1_12
Chen H, Chong W, Yang X, Zhang Y, Sang S, Li X, Lu M (2020) Age-related mutational signature negatively associated with immune activity and survival outcome in triple-negative breast cancer. Oncoimmunology 9:1788252. https://doi.org/10.1080/2162402X.2020.1788252
Chen H, Wang J, Zeng R, Luo Y, Guo K, Wu H, Yang Q, Jiang R, Sha W, Zhuo Z (2021) Development and validation of a novel mitophagy-related gene prognostic signature for hepatocellular carcinoma based on immunoscore classification of tumor. J Oncol 2021:5070099. https://doi.org/10.1155/2021/5070099
Choi KC, Auersperg N, Leung PC (2003) Mitogen-activated protein kinases in normal and (pre)neoplastic ovarian surface epithelium. Reprod Biol Endocrinol 1:71. https://doi.org/10.1186/1477-7827-1-71
Chong W, Wang Z, Shang L et al (2021) Association of clock-like mutational signature with immune checkpoint inhibitor outcome in patients with melanoma and NSCLC. Mol Ther Nucleic Acids 23:89–100. https://doi.org/10.1016/j.omtn.2020.10.033
Colaprico A, Silva TC, Olsen C et al (2016) TCGAbiolinks: an R/bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 44:e71. https://doi.org/10.1093/nar/gkv1507
Dai K, Radin DP, Leonardi D (2019) PINK1 depletion sensitizes non-small cell lung cancer to glycolytic inhibitor 3-bromopyruvate: involvement of ROS and mitophagy. Pharmacol Rep 71:1184–1189. https://doi.org/10.1016/j.pharep.2019.08.002
Dai D, Liu L, Guo Y, Shui Y, Wei Q (2022) A comprehensive analysis of the effects of key mitophagy genes on the progression and prognosis of lung adenocarcinoma. Cancers (Basel). https://doi.org/10.3390/cancers15010057
Davis S, Meltzer PS (2007) GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847. https://doi.org/10.1093/bioinformatics/btm254
Denisenko TV, Gogvadze V, Zhivotovsky B (2021) Mitophagy in carcinogenesis and cancer treatment. Discov Oncol 12:58. https://doi.org/10.1007/s12672-021-00454-1
Ettinger DS, Wood DE, Akerley W et al (2015) Non-small cell lung cancer, version 6.2015. J Natl Compr Canc Netw 13:515–524. https://doi.org/10.6004/jnccn.2015.0071
Goldman MJ, Craft B, Hastie M et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38:675–678. https://doi.org/10.1038/s41587-020-0546-8
Goswami MT, Chen G, Chakravarthi BV et al (2015) Role and regulation of coordinately expressed de novo purine biosynthetic enzymes PPAT and PAICS in lung cancer. Oncotarget 6:23445–23461. https://doi.org/10.18632/oncotarget.4352
Guo JY, Chen HY, Mathew R et al (2011) Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev 25:460–470. https://doi.org/10.1101/gad.2016311
Hanzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform 14:7. https://doi.org/10.1186/1471-2105-14-7
He J, Li X (2022) Identification and validation of aging-related genes in idiopathic pulmonary fibrosis. Front Genet 13:780010. https://doi.org/10.3389/fgene.2022.780010
Hu J, Zhou L, Song Z, Xiong M, Zhang Y, Yang Y, Chen K, Chen Z (2019) The identification of new biomarkers for bladder cancer: a study based on TCGA and GEO datasets. J Cell Physiol 234:15607–15618. https://doi.org/10.1002/jcp.28208
Huang Q, Liu F, Shen J (2019) Bioinformatic validation identifies candidate key genes in diffuse large-B cell lymphoma. Per Med 16:313–323. https://doi.org/10.2217/pme-2018-0068
Hutz K, Zeiler J, Sachs L, Ormanns S, Spindler V (2017) Loss of desmoglein 2 promotes tumorigenic behavior in pancreatic cancer cells. Mol Carcinog 56:1884–1895. https://doi.org/10.1002/mc.22644
Islam T, Rahman R, Gov E, Turanli B, Gulfidan G, Haque A, Arga KY, Haque Mollah N (2018) Drug targeting and biomarkers in head and neck cancers: insights from systems biology analyses. OMICS 22:422–436. https://doi.org/10.1089/omi.2018.0048
Landi MT, Dracheva T, Rotunno M et al (2008) Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS ONE 3:e1651. https://doi.org/10.1371/journal.pone.0001651
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. https://doi.org/10.1016/j.cels.2015.12.004
Liu YF, Yang A, Liu W, Wang C, Wang M, Zhang L, Wang D, Dong JF, Li M (2015) NME2 reduces proliferation, migration and invasion of gastric cancer cells to limit metastasis. PLoS ONE 10:e0115968. https://doi.org/10.1371/journal.pone.0115968
Ma Q, Xu Y, Liao H, Cai Y, Xu L, Xiao D, Liu C, Pu W, Zhong X, Guo X (2019) Identification and validation of key genes associated with non-small-cell lung cancer. J Cell Physiol 234:22742–22752. https://doi.org/10.1002/jcp.28839
Matsuoka R, Yoshida MC, Furutani Y, Imamura S, Kanda N, Yanagisawa M, Masaki T, Takao A (1993) Human smooth muscle myosin heavy chain gene mapped to chromosomal region 16q12. Am J Med Genet 46:61–67. https://doi.org/10.1002/ajmg.1320460110
McCorkle JR, Leonard MK, Kraner SD, Blalock EM, Ma D, Zimmer SG, Kaetzel DM (2014) The metastasis suppressor NME1 regulates expression of genes linked to metastasis and patient outcome in melanoma and breast carcinoma. Cancer Genomics Proteomics 11:175–194
Nencioni L, De Chiara G, Sgarbanti R et al (2009) Bcl-2 expression and p38MAPK activity in cells infected with influenza A virus: impact on virally induced apoptosis and viral replication. J Biol Chem 284:16004–16015. https://doi.org/10.1074/jbc.M900146200
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457. https://doi.org/10.1038/nmeth.3337
Owlanj H, Jie Yang H, Wei Feng Z (2012) Nucleoside diphosphate kinase Nm23-M1 involves in oligodendroglial versus neuronal cell fate decision in vitro. Differentiation 84:281–293. https://doi.org/10.1016/j.diff.2012.08.007
Perkins NJ, Weck J, Mumford SL, Sjaarda LA, Mitchell EM, Pollack AZ, Schisterman EF (2019) Combining biomarker calibration data to reduce measurement error. Epidemiology 30(Suppl 2):S3–S9. https://doi.org/10.1097/EDE.0000000000001094
Peters MJ, Joehanes R, Pilling LC et al (2015) The transcriptional landscape of age in human peripheral blood. Nat Commun 6:8570. https://doi.org/10.1038/ncomms9570
Pollard TD, Weihing RR (1974) Actin and myosin and cell movement. CRC Crit Rev Biochem 2:1–65. https://doi.org/10.3109/10409237409105443
Rahaus M, Desloges N, Wolff MH (2004) Replication of varicella-zoster virus is influenced by the levels of JNK/SAPK and p38/MAPK activation. J Gen Virol 85:3529–3540. https://doi.org/10.1099/vir.0.80347-0
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Rousseaux S, Debernardi A, Jacquiau B et al (2013) Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med 5:186ra166. https://doi.org/10.1126/scitranslmed.3005723
Schonle A, Hartl FA, Mentzel J et al (2016) Caveolin-1 regulates TCR signal strength and regulatory T-cell differentiation into alloreactive T cells. Blood 127:1930–1939. https://doi.org/10.1182/blood-2015-09-672428
Sharma A, Ahmad S, Ahmad T, Ali S, Syed MA (2021) Mitochondrial dynamics and mitophagy in lung disorders. Life Sci 284:119876. https://doi.org/10.1016/j.lfs.2021.119876
Si X, Luo H, Morgan A, Zhang J, Wong J, Yuan J, Esfandiarei M, Gao G, Cheung C, McManus BM (2005) Stress-activated protein kinases are involved in coxsackievirus B3 viral progeny release. J Virol 79:13875–13881. https://doi.org/10.1128/JVI.79.22.13875-13881.2005
Siegel R, DeSantis C, Virgo K et al (2012) Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 62:220–241. https://doi.org/10.3322/caac.21149
Stelzer G, Rosen N, Plaschkes I et al (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform 54:13031–313033. https://doi.org/10.1002/cpbi.5
Stutvoet TS, Kol A, de Vries EG, de Bruyn M, Fehrmann RS, Terwisscha van Scheltinga AG, de Jong S (2019) MAPK pathway activity plays a key role in PD-L1 expression of lung adenocarcinoma cells. J Pathol 249:52–64. https://doi.org/10.1002/path.5280
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. https://doi.org/10.1073/pnas.0506580102
Tataranni T, Piccoli C (2019) Dichloroacetate (DCA) and cancer: an overview towards clinical applications. Oxid Med Cell Longev 2019:8201079. https://doi.org/10.1155/2019/8201079
Trimmer C, Bonuccelli G, Katiyar S, Sotgia F, Pestell RG, Lisanti MP, Capozza F (2013) Cav1 suppresses tumor growth and metastasis in a murine model of cutaneous SCC through modulation of MAPK/AP-1 activation. Am J Pathol 182:992–1004. https://doi.org/10.1016/j.ajpath.2012.11.008
Van Calster B, Wynants L, Verbeek JFM, Verbakel JY, Christodoulou E, Vickers AJ, Roobol MJ, Steyerberg EW (2018) Reporting and interpreting decision curve analysis: a guide for investigators. Eur Urol 74:796–804. https://doi.org/10.1016/j.eururo.2018.08.038
Wang HQ, Yu XD, Liu ZH, Cheng X, Samartzis D, Jia LT, Wu SX, Huang J, Chen J, Luo ZJ (2011) Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J Pathol 225:232–242. https://doi.org/10.1002/path.2931
Wang RJ, Wu P, Cai GX, Wang ZM, Xu Y, Peng JJ, Sheng WQ, Lu HF, Cai SJ (2014) Down-regulated MYH11 expression correlates with poor prognosis in stage II and III colorectal cancer. Asian Pac J Cancer Prev 15:7223–7228. https://doi.org/10.7314/apjcp.2014.15.17.7223
Wang Y, Liu HH, Cao YT, Zhang LL, Huang F, Yi C (2020) The role of mitochondrial dynamics and mitophagy in carcinogenesis, metastasis and therapy. Front Cell Dev Biol 8:413. https://doi.org/10.3389/fcell.2020.00413
Wang Y, Wang Z, Sun J, Qian Y (2021) Identification of HCC subtypes with different prognosis and metabolic patterns based on mitophagy. Front Cell Dev Biol 9:799507. https://doi.org/10.3389/fcell.2021.799507
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Zeng X, Xu C, Cheng J, Sun C, Wang Z, Gong Z, Long H, Zhu B (2020) Poor glycemic control might compromise the efficacy of chemotherapy in non-small cell lung cancer patients with diabetes mellitus. Cancer Med 9:902–911. https://doi.org/10.1002/cam4.2750
Zhang W, Li Y, Lyu J, Shi F, Kong Y, Sheng C, Wang S, Wang Q (2022) An aging-related signature predicts favorable outcome and immunogenicity in lung adenocarcinoma. Cancer Sci 113:891–903. https://doi.org/10.1111/cas.15254
Zhou X, Cai L, Liu J, Hua X, Zhang Y, Zhao H, Wang B, Li B, Gai P (2018) Analyzing EGFR mutations and their association with clinicopathological characteristics and prognosis of patients with lung adenocarcinoma. Oncol Lett 16:362–370. https://doi.org/10.3892/ol.2018.8681
Zhou S, Yan Y, Chen X et al (2019) Roles of highly expressed PAICS in lung adenocarcinoma. Gene 692:1–8. https://doi.org/10.1016/j.gene.2018.12.064
Zhuo Z, Lin H, Liang J, Ma P, Li J, Huang L, Chen L, Yang H, Bai Y, Sha W (2021) Mitophagy-related gene signature for prediction prognosis, immune scenery, mutation, and chemotherapy response in pancreatic cancer. Front Cell Dev Biol 9:802528. https://doi.org/10.3389/fcell.2021.802528
Acknowledgements
This work was supported by National Key R&D Program of China (Grant Nos. 2020YFE02022200).
Funding
This work was supported by National Key R&D Program of China (Grant no. 2020YFE02022200).
Author information
Authors and Affiliations
Contributions
Data collection: XM, WS; Data analysis: XM, WS, ML; Manuscript writing: XM, BZ; Study design: YG, XM.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
No ethics approval should be declared.
Consent to participate
This study does not cover this content.
Consent to publish
This study does not cover this content.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
432_2023_5390_MOESM1_ESM.tif
Supplementary file1Fig. S1 TCGA-LUAD dataset normalization. (a-b) Boxplot of TCGA-LUAD dataset pre (a) and post (b) normalization. (c-d) Principal component analysis plot of TCGA-LUAD dataset pre (c) and post (d) normalization (TIF 3930 KB)
432_2023_5390_MOESM2_ESM.tif
Supplementary file2Fig. S2 Normalization of the GEO LUAD datasets. (a-b) Boxplot of the GSE10072 dataset before (A) and after (B) normalization. (c-d) Boxplot of the GSE30219 dataset pre (c) and post (d) normalization (TIF 3234 KB)
432_2023_5390_MOESM3_ESM.tif
Supplementary file3Fig. S3 Correlation analysis of mitophagy-aging differentially expressed genes (MiAg DEGs) in lung adenocarcinoma (LUAD) in the (a) GSE30219 and (d) GSE10072dataset. (a) Correlation heat map of MiAg DEGs. Correlation scatter plots of MiAg DEGs MYH11 and NME1 (b-c) and NME1 and PAICS (e-f). ns, not significant (P ≥ 0.05); *P < 0.05; **P < 0.01; ***P < 0.001. In a correlation scatter diagram, the strength of the correlation coefficient (r) is determined by its absolute value. Absolute value > 0.8: strong correlation; 0.5–0.8: moderately correlated; 0.3–0.5: weak correlation; <0.3 weak and irrelevant correlation (TIF 718 KB)
432_2023_5390_MOESM4_ESM.tif
Supplementary file4Fig. S4 Differential expression analysis of mitophagy-aging differentially expressed genes (MiAg DEGs) in the GSE10072 dataset. (a) Group comparison chart of the differential expression analysis of MiAg DEGs. (b–h) Receiver operating characteristic curve of MiAg DEGs CAV1 (b), DSG2 (c), DSP (d), MYH11 (e), NME1 (f), PAICS (g), PLOD2 (h). ns, not significant (P ≥ 0.05); *P < 0.05; **P < 0.01; ***P < 0.001. The closer the area under the curve (AUC) is to 1, the better the diagnostic effect. AUC = 0.5–0.7: low accuracy; 0.7–0.9: certain accuracy; >0.9: high accuracy (TIF 939 KB)
432_2023_5390_MOESM5_ESM.tif
Supplementary file5Fig. S5 Differential expression analysis of mitophagy-aging-related differentially expressed genes (MiAg DEGs) in the GSE30219 dataset. (a) Group comparison chart of the differential expression analysis of MiAg DEGs. (b–h) Receiver operating characteristic curve results of MiAg DEGs CAV1 (b), DSG2 (c), DSP (d), MYH11 (e), NME1 (f), PAICS (g), PLOD2 (h). ns, not significant (P ≥ 0.05); *P < 0.05; **P < 0.01; ***P < 0.001. The closer the area under the curve (AUC) value is to 1, the better the diagnostic effect. AUC = 0.5–0.7: low accuracy; 0.7–0.9: certain accuracy; > 0.9: high accuracy (TIF 935 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Meng, X., Song, W., Zhou, B. et al. Prognostic and immune correlation analysis of mitochondrial autophagy and aging-related genes in lung adenocarcinoma. J Cancer Res Clin Oncol 149, 16311–16335 (2023). https://doi.org/10.1007/s00432-023-05390-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05390-x