Abstract
Purpose
Prognostic prediction is a challenging task in cytogenetically normal acute myeloid leukemia (CN-AML) patients. In this study, we aimed at developing a novel prognostic signature to predict and stratify the survival of CN-AML patients.
Methods
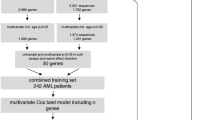
Using a training dataset (GSE12417), 5-gene prognostic signature was established to predict survival of CN-AML patients. The prognostic performance of this prognostic signature was further validated in testing dataset (TCGA CN-AML cohort) and validation dataset (GSE6891 CN-AML cohort).
Results
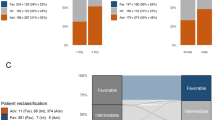
In training, testing and validation datasets, the increased 5-gene risk score was significantly related with inferior overall survival (OS) of patients, and the area under the receiver operating characteristic curve (AUC) demonstrated that our prognostic signature had overall prediction accuracy. The excellent prognostic value of the 5-gene prognostic signature was also supported by the comparison with three previously proposed prognostic models. For the intermediate-risk CN-AML patients and the CN-AML patients with FLT3 or NPM1 mutation, our model could also well dichotomize them into two subgroups with distinct prognosis. Multivariate analysis demonstrated that 5-gene risk score was the only independent risk factor in TCGA CN-AML cohort. Nomogram including the 5-gene risk score performed well in predicting 1-year, 2-year and 3-year OS.
Conclusion
In summary, our novel 5-gene prognostic signature facilitated the improvement in risk stratification of CN-AML patients.





Similar content being viewed by others
Data availability
All gene expression profile data and corresponding clinical information of CN-AML patients are available in Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/).
References
Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al (2016) The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 127(20):2391–2405
Azad AK, Rajaram MV, Schlesinger LS (2014) Exploitation of the macrophage mannose receptor (CD206) in infectious disease diagnostics and therapeutics. J Cytol Mol Biol 1(1):1000003
Chen Y, Pacyna-Gengelbach M, Deutschmann N, Niesporek S, Petersen I (2007) Homeobox gene HOP has a potential tumor suppressive activity in human lung cancer. Int J Cancer 121(5):1021–1027
Daver N, Schlenk RF, Russell NH, Levis MJ (2019) Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia 33(2):299–312
Döhner K, Schlenk RF, Habdank M, Scholl C, Rücker FG, Corbacioglu A et al (2005) Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood 106(12):3740–3746
Döhner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Büchner T et al (2017) Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 129(4):424–447
Doorn-Khosrovani SBW, Erpelinck C, Meijer J, Oosterhoud S, Putten WL, Valk PJM et al (2003) Biallelic mutations in the CEBPA gene and low CEBPA expression levels as prognostic markers in intermediate-risk AML. Hematol J 4(1):31–40
Fu L, Fu HP, Tian L, Xu KM, Hu K, Wang J et al (2016) High expression of RUNX1 is associated with poorer outcomes in cytogenetically normal acute myeloid leukemia. Oncotarget 7(13):15828–15839
Hou HA, Tien HF (2020) Genomic landscape in acute myeloid leukemia and its implications in risk classification and targeted therapies. J Biomed Sci 27(1):81
Katoh H, Yamashita K, Waraya M, Margalit O, Ooki A, Tamaki H et al (2012) Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia 14(7):559–571
Komanduri KV, Levine RL (2016) Diagnosis and therapy of acute myeloid leukemia in the era of molecular risk stratification. Annu Rev Med 67:59–72
Li Z, Herold T, He C, Valk PJ, Chen P, Jurinovic V et al (2013) Identification of a 24-gene prognostic signature that improves the European LeukemiaNet risk classification of acute myeloid leukemia: an international collaborative study. J Clin Oncol 31(9):1172–1181
Lin CC, Hsu YC, Li YH, Kuo YY, Hou HA, Lan KH et al (2017) Higher HOPX expression is associated with distinct clinical and biological features and predicts poor prognosis in de novo acute myeloid leukemia. Haematologica 102(6):1044–1053
Liu Y, Zhang W (2020) The role of HOPX in normal tissues and tumor progression. Biosci Rep 40(1):BSR20191953
Liu K, Lai M, Wang S, Zheng K, Xie S, Wang X (2020) Construction of a CXC chemokine-based prediction model for the prognosis of colon cancer. Biomed Res Int 2020:6107865
Ma QL, Wang JH, Wang YG, Hu C, Mu QT, Yu MX et al (2015) High IDH1 expression is associated with a poor prognosis in cytogenetically normal acute myeloid leukemia. Int J Cancer 137(5):1058–1065
Ma QL, Wang JH, Yang M, Wang HP, Jin J (2018) MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Transl Med 16(1):68
Mendler JH, Maharry K, Radmacher MD, Mrozek K, Becker H, Metzeler KH et al (2012) RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and MicroRNA expression signatures. J Clin Oncol 30(25):3109–3118
Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N et al (2016) A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540(7633):433–437
Qu X, Othus M, Davison J, Wu Y, Yan L, Meshinchi S et al (2017) Prognostic methylation markers for overall survival in cytogenetically normal patients with acute myeloid leukemia treated on SWOG trials. Cancer 123(13):2472–2481
Ruan GT, Gong YZ, Liao XW, Wang S, Huang W, Wang XK et al (2019) Diagnostic and prognostic values of CXC motif chemokine ligand 3 in patients with colon cancer. Oncol Rep 42(5):1996–2008
Shang J, Song Q, Yang ZY, Li DY, Chen WJ, Luo L et al (2017) Identification of lung adenocarcinoma specific dysregulated genes with diagnostic and prognostic value across 27 TCGA. Oncotarget 8(50):87292–87306
Shi JL, Fu L, Ang Q, Wang GJ, Zhu J, Wang WD (2015) Overexpression of ATP1B1 predicts an adverse prognosis in CN-AML. Oncotarget 7(3):2585–2595
Song Y, Zhang W, He X, Liu X, Yang P, Wang J et al (2019) High NCALD expression predicts poor prognosis of cytogenetic normal acute myeloid leukemia. J Transl Med 17(1):166
Torres CM, Biran A, Burney MJ, Patel H, Henser-Brownhill T, Cohen AS et al (2016) The linker histone H10 generates epigenetic and functional intratumor heterogeneity. Science 353(2307):aaf1644
Wang M, Lindberg J, Klevebring D, Nilsson C, Lehmann S, Gronberg H et al (2018) Development and validation of a novel RNA sequencing-based prognostic score for acute myeloid leukemia. J Natl Cancer Inst 110(10):1094–1101
Wu J, Liu XJ, Hu JN, Liao XH, Lin FF (2020) Transcriptomics and prognosis analysis to identify critical biomarkers in invasive breast carcinoma. Technol Cancer Res Treat 19:1533033820957011
Xie F, He M, He L, Liu K, Li M, Hu G et al (2017) Bipartite network analysis reveals metabolic gene expression profiles that are highly associated with the clinical outcomes of acute myeloid leukemia. Comput Biol Chem 67:150–157
Xu ZJ, Gu Y, Wang CZ, Jin Y, Wen XM, Ma JC et al (2020) The M2 macrophage marker CD206: a novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 9(1):1683347
Yamaguchi S, Asanoma K, Takao T, Kato K, Wake N (2009) Homeobox gene HOPX is epigenetically silenced in human uterine endometrial cancer and suppresses estrogen-stimulated proliferation of cancer cells by inhibiting serum response factor. Int J Cancer 124(11):2577–2588
Yang Z, Shang J, Li N, Zhang L, Tang T, Tian G et al (2020) Development and validation of a 10-gene prognostic signature for acute myeloid leukaemia. J Cell Mol Med 24(8):4510–4523
Zhou JD, Yao DM, Han L, Xiao GF, Guo H, Zhang TJ et al (2017) Low NKD1 expression predicts adverse prognosis in cytogenetically normal acute myeloid leukemia. Tumour Biol 39(4):1010428317699123
Funding
This study was funded by Xinjiang Joint Fund of National Natural Science Foundation of China (U1903117).
Author information
Authors and Affiliations
Contributions
CD, TZ, LF designed the study. CD, PZ, ZH performed the study. CD, WH wrote the manuscript. SZ, WZ, XH analyzed clinical and molecular data and performed statistical analysis. TZ, LF provided critical revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
All gene expression profile data and corresponding clinical information of CN-AML patients were downloaded from open-Access databases, so the ethical declaration is not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deng, C., Zeng, T., Zhu, P. et al. A novel 5-gene prognostic signature to improve risk stratification of cytogenetically normal acute myeloid leukemia. J Cancer Res Clin Oncol 149, 10015–10025 (2023). https://doi.org/10.1007/s00432-023-04884-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04884-y