Abstract
Purpose
Esophageal squamous cell carcinoma (ESCC), is a frequent digestive tract malignant carcinoma with a high fatality rate. Daphne altaica (D. altaica), a medicinal plant that is frequently employed in Kazakh traditional medicine, and which has traditionally been used to cure cancer and respiratory conditions, but research on the mechanism is lacking. Therefore, we examined and verified the hub genes and mechanism of D. altaica treating ESCC.
Methods
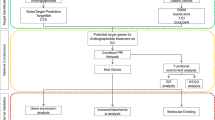
Active compounds and targets of D. altaica were screened by databases such as TCMSP, and ESCC targets were screened by databases such as GeneCards and constructed the compound-target network and PPI network. Meantime, data sets between tissues and adjacent non-cancerous tissues from GEO database (GSE100942, GPL570) were analyzed to obtain DEGs using the limma package in R. Hub genes were validated using data from the Kaplan–Meier plotter database, TIMER2.0 and GEPIA2 databases. Finally, AutoDock software was used to predict the binding sites through molecular docking.
Results
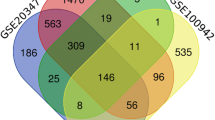
In total, 830 compound targets were obtained from TCMSP and other databases. In addition, 17,710 disease targets were acquired based on GeneCards and other databases. In addition, we constructed the compound-target network and PPI network. Then, 127 DEGs were observed (82 up-regulated and 45 down-regulated genes). Hub genes were screened including TOP2A, NUF2, CDKN2A, BCHE, and NEK2, and had been validated with the help of several publicly available databases. Finally, molecular docking results showed more stable binding between five hub genes and active compounds.
Conclusions
In the present study, five hub genes were screened and validated, and potential mechanisms of action were predicted, which could provide a theoretical understanding of the treatment of ESCC with D. altaica.






Similar content being viewed by others
Data availability
The datasets (GSE100942, GPL570) analysed during the current study are available in the [The Gene Expression Omnibus (GEO) database] repository, [https://www.ncbi.nlm.nih.gov/geo/].
References
Abid R, Ghazanfar S, Farid A, Sulaman SM, Idrees M, Amen RA, Muzammal M, Shahzad MK, Mohamed MO, Khaled AA, Safir W, Ghori I, Elasbali AM, Alharbi B (2022) Pharmacological properties of 4’, 5, 7-trihydroxyflavone (Apigenin) and its impact on cell signaling pathways. Molecules 27(13):4304. https://doi.org/10.3390/molecules27134304
Agostini M, Melino G, Bernassola F (2018) The p53 family in brain disease. Antioxid Redox Signal 29(1):1–14. https://doi.org/10.1089/ars.2017.7302
Alsop BR, Sharma P (2016) Esophageal cancer. Gastroenterol Clin N Am 45:399–412. https://doi.org/10.1016/j.gtc.2016.04.001
Alwhaibi A, Verma AMS, Somanath PR (2019) The unconventional role of Akt1 in the advanced cancers and in diabetes-promoted carcinogenesis. Pharmacol Res 145:104270. https://doi.org/10.1016/j.phrs.2019.104270
Badawy OM, Loay I (2019) FISH analysis of TOP2A and HER-2 aberrations in female breast carcinoma on archived material: Egyptian NCI experience. Appl Immunohistochem Mol Morphol. https://doi.org/10.1097/PAI.0000000000000574
Berman HM, Westbrook J, Feng Z et al (2000) The protein data bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Burgering BM, Coffer PJ (1995) Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature 376:599–602. https://doi.org/10.1038/376599a0
Chen P, Zhang JY, Sha BB, Ma YE, Hu T, Ma YC, Sun H, Shi JX, Dong ZM, Li P (2017) Luteolin inhibits cell proliferation and induces cell apoptosis via down-regulation of mitochondrial membrane potential in esophageal carcinoma cells EC1 and KYSE450. Oncotarget 8(16):27471–27480. https://doi.org/10.18632/oncotarget.15832
Chen Z, Guo Y, Zhao D, Zou Q, Yu F, Zhang L, Xu L (2021) Comprehensive analysis revealed that CDKN2A is a biomarker for immune infiltrates in multiple cancers. Front Cell Dev Biol 9:808208. https://doi.org/10.3389/fcell.2021.808208
Chen XC, Pang LJ, Li F (2006) Progress in the study of esophageal cancer in Kazakhs in Xinjiang. J Nongken Med 28(5): 384–387. https://kns.cnki.net/kcms/detail/detail
Conceição AL, Babeto E, Candido NM, Franco FC, de Campos Zuccari DA, Bonilha JL, Cordeiro JA, Calmon MF, Rahal P (2015) Differential expression of ADAM23, CDKN2A (P16), MMP14 and VIM associated with giant cell tumor of bone. J Cancer 6(7):593–603. https://doi.org/10.7150/jca.11238
Cong J (2015) Research on the chemical constituents of the chloroform extraction layer of Daphne Altaica Pall. Northeast Normal University https://kns.cnki.net/KCMS/detail/detail
de Resende MF, Vieira S, Chinen LT et al (2013) Prognostication of prostate cancer based on TOP2A protein and gene assessment: TOP2A in prostate cancer. J Transl Med 11:36. https://doi.org/10.1186/1479-5876-11-36
Du X, Xue Z, Lv J, Wang H (2020) Expression of the topoisomerase II alpha (TOP2A) gene in lung adenocarcinoma cells and the association with patient outcomes. Med Sci Monit 26:e929120. https://doi.org/10.12659/MSM.929120
Fang Y, Kong Y, Xi J, Zhu M, Zhu T, Jiang T, Hu W, Ma M, Zhang X (2016) Preclinical activity of MBM-5 in gastrointestinal cancer by inhibiting NEK2 kinase activity. Oncotarget 7:79327–79341
Foulkes WD, Flanders TY, Pollock PM, Hayward NK (1997) The CDKN2A (p16) gene and human cancer. Mol Med 3:5–20
Han RH (2015) China Kazakh and Kazakhstan Kazakh cultural comparison diet. Xinjiang Normal University, Urumqi. https://kns.cnki.net/kcms/detail/detail
Hsin KY, Ghosh S, Kitano H (2013) Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One 8:e83922
Johnson G, Moore SW (2013) Why has butyrylcholinesterase been retained? Structural and functional diversification in a duplicated gene. Neurochem Int 61:783–797. https://doi.org/10.1016/j.neuint.2012.06.016
Kastenhuber ER, Lowe SW (2017) Putting p53 in context. Cell 170(6):1062–1078. https://doi.org/10.1016/j.cell.2017.08.028
Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, Li Q, Shoemaker BA, Thiessen PA, Yu B, Zaslavsky L, Zhang J, Bolton EE (2019) PubChem 2019 update: improved access to chemical data. Nucleic Acids Res 47:D1102-d1109
Kizaibek M, Daniar M, Li L, Upur H (2011) Antiproliferative activity of different extracts from Daphne altaica Pall. On selected cancer cells. J Med Plants Res 5:3448–3452
Kizaibek M, Wubuli A, Gu Z et al (2020) Effects of an ethyl acetate extract of Daphne altaica stem bark on the cell cycle, apoptosis and expression of PPARγ in Eca-109 human esophageal carcinoma cells. Mol Med Rep 22(2):1400–1408. https://doi.org/10.3892/mmr.2020.11187
Lampón N, Hermida-Cadahia EF, Riveiro A, Tutor JC (2012) Association between butyrylcholinesterase activity and low-grade systemic inflammation. Ann Hepatol 11(3):356–363. https://doi.org/10.1016/S1665-2681(19)30932-9
Li YL, Min W, Zhang W (2009) Influence of network topology on critical coupling parameters in synchronization of small world neutral networks. Far East J Dyn Syst 11(1):65–75
Liu Z, Sun Y, Zhen H, Nie C (2022a) Network pharmacology integrated with transcriptomics deciphered the potential mechanism of Codonopsis pilosula against hepatocellular carcinoma. Evid Based Complement Altern Med 2022:1340194. https://doi.org/10.1155/2022/1340194
Liu J, Tian T, Liu X, Cui Z (2022b) BCHE as a prognostic biomarker in endometrial cancer and its correlation with immunity. J Immunol Res 2022:6051092. https://doi.org/10.1155/2022/6051092
Lockridge O (2015) Review of human butyrylcholinesterase structure, function, genetic variants, history of use in the clinic, and potential therapeutic uses. Pharmacology 148:34–46. https://doi.org/10.1016/j.pharmthera.2014.11.011
Macheret M, Halazonetis TD (2015) DNA replication stress as a hallmark of cancer. Annu Rev Pathol 10:425–448. https://doi.org/10.1146/annurev-pathol-012414-040424
Manning BD, Toker A (2017) AKT/PKB signaling: navigating the network. Cell 169:381–405. https://doi.org/10.1016/j.cell.2017.04.001
McKenna M, Balasuriya N, Zhong S, Li SS, O’Donoghue P (2021) Phospho-form specific substrates of protein kinase B (AKT1). Front Bioeng Biotechnol 8:619252. https://doi.org/10.3389/fbioe.2020.619252
Morris GM, Huey R, Lindstrom W et al (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. https://doi.org/10.1002/jcc.21256
Murat K, Xamxinur Y, Su JC (2016) Progress in the traditional application and anticancer activity of Kazakh medicine Daphne altaica and its congeners. Hebei Med J 38(19):3007–3010. https://kns.cnki.net/kcms/detail/detail
Nagy A, Munkacsy G, Gyorffy B (2021) Pancancer survival analysis of cancer hallmark genes. Sci Rep 11(1):6047. https://doi.org/10.1038/s41598-021-84787-5
Nakajima M, Kato H (2013) Treatment options for esophageal squamous cell carcinoma. Expert Opin Pharmacother 14:1345–1354. https://doi.org/10.1517/14656566.2013.801454
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminf 3:33
Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M (1999) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27(1):29–34. https://doi.org/10.1093/nar/27.1.29
Ou R, Mo L, Tang H, Leng S, Zhu H, Zhao L, Ren Y, Xu Y (2020) circRNA-AKT1 sequesters miR-942-5p to upregulate AKT1 and promote cervical cancer progression. Mol Ther Nucleic Acids 20:308–322. https://doi.org/10.1016/j.omtn.2020.01.003
Rahmani AH, Alsahli MA, Almatroudi A, Almogbel MA, Khan AA, Anwar S, Almatroodi SA (2022) The potential role of apigenin in cancer prevention and treatment. Molecules 27(18):6051. https://doi.org/10.3390/molecules27186051
Robles AI, Harris CC (2010) Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol 2(3):a001016. https://doi.org/10.1101/cshperspect.a001016
Shi L (2015) Research on the chemical constituents of ethyl acetate extract from Daphne altaica. Northeast Normal University. https://kns.cnki.net/KCMS/detail/detail
Su W, Hu H, Ding Q, Wang M, Zhu Y, Zhang Z, Geng Z, Lin S, Zhou P (2022) NEK2 promotes the migration and proliferation of ESCC via stabilization of YAP1 by phosphorylation at Thr-143. Cell Commun Signal 20(1):87. https://doi.org/10.1186/s12964-022-00898-0
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
The Gene Ontology Consortium (2017) Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45(D1):D331–D338. https://doi.org/10.1093/nar/gkw1108
Tungekar A, Mandarthi S, Mandaviya PR et al (2018) ESCC ATLAS: a population wide compendium of biomarkers for esophageal squamous cell carcinoma. Sci Rep 8(1):12715. https://doi.org/10.1038/s41598-018-30579-3
Tustumi F, Kimura MS, Takeda FR, Uema RH, Salum RA, Ribeiro-Junior U, Cecconello I (2016) Prognostic factors and survival analysis in esophageal carcinoma. Arq Bras Cir Dig 29:138–141. https://doi.org/10.1590/0102-6720201600030003
Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137(3):413–431. https://doi.org/10.1016/j.cell.2009.04.037
Walters WP, Murcko MA (2002) Prediction of “drug-likeness.” Adv Drug Deliv Rev 54(3):255–271. https://doi.org/10.1016/s0169-409x(02)00003-0
Wang M, Firrman J, Liu L, Yam K (2019) A review on flavonoid apigenin: dietary intake, ADME, antimicrobial effects, and interactions with human gut microbiota. Biomed Res Int 2019:7010467. https://doi.org/10.1155/2019/7010467
Wang T, Lu J, Wang R, Cao W, Xu J (2022) TOP2A promotes proliferation and metastasis of hepatocellular carcinoma regulated by miR-144-3p. J Cancer 13(2):589–601. https://doi.org/10.7150/jca.64017
Xi JB, Fang YF, Frett B, Zhu ML, Zhu T, Kong YN, Guan FJ, Zhao Y, Zhang XW, Li HY, Ma ML, Hu W (2017) Structure-based design and synthesis of imidazo[1,2-a] pyridine derivatives as novel and potent Nek2 inhibitors with in vitro and in vivo antitumor activities. Eur J Med Chem 126:1083–1106
Xia J, Franqui-Machin R, Gu Z, Zhan F (2015) Role of NEK2A in human cancer and its therapeutic potentials. Biomed Res Int 2015:12
Xie X, Jiang S, Li X (2021) Nuf2 is a prognostic-related biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Front Oncol 11:621373. https://doi.org/10.3389/fonc.2021.621373
Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, Duan J, Ling Y (2012) A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 13(6):6964–6982. https://doi.org/10.3390/ijms13066964
Yang W, Zhao X, Han Y, Duan L, Lu X, Wang X, Zhang Y, Zhou W, Liu J, Zhang H, Zhao Q, Hong L, Fan D (2019) Identification of hub genes and therapeutic drugs in esophageal squamous cell carcinoma based on integrated bioinformatics strategy. Cancer Cell Int 19:142. https://doi.org/10.1186/s12935-019-0854-6
Yao L, Zhong X, Huang G, Ma Q, Xu L, Xiao H, Guo X (2021) Investigation on the potential correlation between TP53 and esophageal cancer. Front Cell Dev Biol 9:730337. https://doi.org/10.3389/fcell.2021.730337
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Zeng S, Liu A, Dai L et al (2019) Prognostic value of TOP2A in bladder urothelial carcinoma and potential molecular mechanisms. BMC Cancer 19(1):604. https://doi.org/10.1186/s12885-019-5814-y
Zhang R, Xu J, Zhao J, Bai JH (2018) Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J Cell Biochem 119(9):7256–7263. https://doi.org/10.1002/jcb.26916
Zhang HX, Chen Y, Yin D et al (2009) Exploration of the risk factors of esophageal cancer in Xinjiang Kazakh nationality. Mod Prev Med 36(10): 1804–1806. https://kns.cnki.net/kcms/detail/detail
Zheng S, Vuitton L, Sheyhidin I, Vuitton DA, Zhang Y, Lu X (2010) Northwestern China: a place to learn more on oesophageal cancer. Part one: behavioural and environmental risk factors. Eur J Gastroenterol Hepatol 22(8):917–925. https://doi.org/10.1097/MEG.0b013e3283313d8b
Zheng S, Vuitton L, Sheyhidin I, Vuitton DA, Zhang Y, Lu X (2011) Northwestern China: a place to learn more on oesophageal cancer. Part two: gene alterations and polymorphisms. Eur J Gastroenterol Hepatol 23(12):1087–1099. https://doi.org/10.1097/MEG.0b013e32834a14d9
Zheng L, Li L, Xie J, Jin H, Zhu N (2021) Six novel biomarkers for diagnosis and prognosis of esophageal squamous cell carcinoma: validated by scRNA-seq and qPCR. J Cancer 12(3):899–911. https://doi.org/10.7150/jca.50443
Zhu H, Jin H, Pi J, Bai H, Yang F, Wu C, Jiang J, Cai J (2016) Apigenin induced apoptosis in esophageal carcinoma cells by destruction membrane structures. Scanning 38(4):322–328. https://doi.org/10.1002/sca.21273
Acknowledgements
We are grateful to all the researchers who provided the data we used.
Funding
The authors appreciate financial support from the funding of Tianshan Talents-Youth Science and Technology Innovation Talents Training Program of Xinjiang Autonomous Region (2022TSYCCX0035); National Natural Science Foundation of China (81660696); "Fourteenth Five-Year Plan" Key Discipline Construction Project of Xinjiang Autonomous Region (2021); The key Laboratory of Xinjiang Autonomous Region (XJDX1713).
Author information
Authors and Affiliations
Contributions
SH, ZT, and WZ designed the study. SH, KA, MH performed bioinformatic analysis. JS, DD, and NN contributed to the conception of the study and drafted the manuscript. MN, NK, and NM contributed to the writing of the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hailati, S., Talihati, Z., Abudurousuli, K. et al. Exploring the hub genes and mechanisms of Daphne altaica treating esophageal squamous cell carcinoma based on network pharmacology and bioinformatics analysis. J Cancer Res Clin Oncol 149, 8467–8481 (2023). https://doi.org/10.1007/s00432-023-04797-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-04797-w