Abstract
Aims
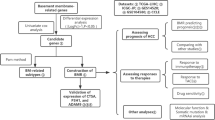
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer. Expression defects and turnover of basement membrane (BM) proteins are key pathogenic factors in cancer. It is still uncertain how the expression of BM-related genes (BMGs) in HCC relates to prognosis.
Methods
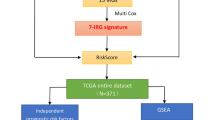
All of the HCC cohort's RNA-seq and clinical information came from TCGA datasets. The least absolute shrinkage and selection operator (LASSO) regression algorithm was utilized to filter down the candidate genes and construct the prognostic model. Univariate and multivariate Cox analyses were run to examine if the risk score may serve as a standalone prognostic indicator. The single-sample gene set enrichment analysis (ssGSEA) was utilized to analyze examine immune cell infiltration and pathway activity.
Results
Five genes and their risk coefficients were eventually identified and patients with HCC were classified as either high or low risk based on the median of risk scores. Multivariate Cox regression analysis found a significant correlation between risk score and OS (p < 0.001). Subgroup analysis showed that BMGs signature had good prediction ability for HCC patients in age, gender, T stage, and AJCC stage (all p < 0.05). According to the ssGSEA, the high-risk subgroup showed higher levels of immune cell infiltration and immune-related pathways were more engaged in the high-risk group.
Conclusions
Our research systematically built a prognostic model using risk score based on BMGs signature in HCC patients. The immune feature analysis of the BMGs signature indicated a potential regulation between tumor immunity and BM in HCC.






Similar content being viewed by others
Data availability
Our data are from the analysis of articles and can be provided when necessary.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- BM:
-
Basement membrane
- BMGs:
-
BM-related genes
- LASSO:
-
Least absolute shrinkage and selection operator
- ssGSEA:
-
Single-sample gene set enrichment analysis
- TKIs:
-
Tyrosine kinase inhibitors
- ICIs:
-
Immune checkpoint inhibitors
- ECM:
-
Extracellular matrix
- TME:
-
Tumor microenvironment
- PPI:
-
Protein–protein interaction
- GO:
-
Gene Ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DEGs:
-
Differentially expressed genes
- CTSA:
-
Cathepsin A
References
Aumailley M (2014) The laminin family. Cell Adh Migr 7(1):48–55. https://doi.org/10.4161/cam.22826
Caciotti A, Catarzi S, Tonin R et al (2013) Galactosialidosis: review and analysis of CTSA gene mutations. Orphanet J Rare Dis 8:114. https://doi.org/10.1186/1750-1172-8-114
Chang TT, Thakar D, Weaver VM (2017) Force-dependent breaching of the basement membrane. Matrix Biol 57–58:178–189. https://doi.org/10.1016/j.matbio.2016.12.005
Clément B, Rescan PY, Baffet G et al (1988) Hepatocytes may produce laminin in fibrotic liver and in primary culture. Hepatology 8(4):794–803. https://doi.org/10.1002/hep.1840080417
Cretu A, Brooks PC (2007) Impact of the non-cellular tumor microenvironment on metastasis: potential therapeutic and imaging opportunities. J Cell Physiol 213(2):391–402. https://doi.org/10.1002/jcp.21222
Deng M, Sun S, Zhao R et al (2022) The pyroptosis-related gene signature predicts prognosis and indicates immune activity in hepatocellular carcinoma. Mol Med 28(1):16. https://doi.org/10.1186/s10020-022-00445-0
Hou G, Li X, Zheng Y et al (2020) Construction and validation of a novel prognostic nomogram for patients with sarcomatoid renal cell carcinoma: a SEER-based study. Int J Clin Oncol 25(7):1356–1363. https://doi.org/10.1007/s10147-020-01681-2
Hu B, Zhu X, Lu J (2020) Cathepsin A knockdown decreases the proliferation and invasion of A549 lung adenocarcinoma cells. Mol Med Rep 21(6):2553–2559. https://doi.org/10.3892/mmr.2020.11068
Jacobetz MA, Chan DS, Neesse A et al (2012) Hyaluronan impairs vascular function and drug delivery in a mouse model of pancreatic cancer. Gut 62(1):112–120. https://doi.org/10.1136/gutjnl-2012-302529
Jayadev R, Morais M, Ellingford JM et al (2022) A basement membrane discovery pipeline uncovers network complexity, regulators, and human disease associations. Sci Adv 8(20):eabn2265. https://doi.org/10.1126/sciadv.abn2265
Khalyfa AA, Punatar S, Yarbrough A (2022) Hepatocellular carcinoma: understanding the inflammatory implications of the microbiome. Int J Mol Sci. https://doi.org/10.3390/ijms23158164
Kuang DM, Peng C, Zhao Q, Wu Y, Chen MS, Zheng L (2010) Activated monocytes in peritumoral stroma of hepatocellular carcinoma promote expansion of memory T helper 17 cells. Hepatology 51(1):154–164. https://doi.org/10.1002/hep.23291
Langhans B, Nischalke HD, Krämer B et al (2019) Role of regulatory T cells and checkpoint inhibition in hepatocellular carcinoma. Cancer Immunol Immunother 68(12):2055–2066. https://doi.org/10.1007/s00262-019-02427-4
Lee CK, Chan SL, Chon HJ (2022) Could we predict the response of immune checkpoint inhibitor treatment in hepatocellular carcinoma? Cancers (basel). https://doi.org/10.3390/cancers14133213
Li L, Wei JR, Dong J et al (2021) Laminin γ2-mediating T cell exclusion attenuates response to anti-PD-1 therapy. Sci Adv. https://doi.org/10.1126/sciadv.abc8346
Li H, Wang G, Wang W et al (2021) A focal adhesion-related gene signature predicts prognosis in glioma and correlates with radiation response and immune microenvironment. Front Oncol 11:698278. https://doi.org/10.3389/fonc.2021.698278
Liu Z, Liu X, Liang J et al (2021) Immunotherapy for hepatocellular carcinoma: Current status and future prospects. Front Immunol 12:765101. https://doi.org/10.3389/fimmu.2021.765101
Liu X, Qiao Y, Chen J, Ge G (2022) Basement membrane promotes tumor development by attenuating T cell activation. J Mol Cell Biol. https://doi.org/10.1093/jmcb/mjac006
Lu P, Weaver VM, Werb Z (2012) The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196(4):395–406. https://doi.org/10.1083/jcb.201102147
Määttä M, Virtanen I, Burgeson R, Autio-Harmainen H (2016) Comparative analysis of the distribution of laminin chains in the basement membranes in some malignant epithelial tumors: the alpha1 chain of laminin shows a selected expression pattern in human carcinomas. J Histochem Cytochem 49(6):711–726. https://doi.org/10.1177/002215540104900605
Menard LC, Fischer P, Kakrecha B et al (2018) Renal cell carcinoma (RCC) tumors display large expansion of double positive (DP) CD4+CD8+ T cells with expression of exhaustion markers. Front Immunol 9:2728. https://doi.org/10.3389/fimmu.2018.02728
Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO (2014) Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 14:518. https://doi.org/10.1186/1471-2407-14-518
Oura K, Morishita A, Tani J, Masaki T (2021) Tumor immune microenvironment and immunosuppressive therapy in hepatocellular carcinoma: a review. Int J Mol Sci. https://doi.org/10.3390/ijms22115801
Park S, Kwon W, Park JK et al (2020) Suppression of cathepsin a inhibits growth, migration, and invasion by inhibiting the p38 MAPK signaling pathway in prostate cancer. Arch Biochem Biophys 688:108407. https://doi.org/10.1016/j.abb.2020.108407
Petz M, Them NC, Huber H, Mikulits W (2012) PDGF enhances IRES-mediated translation of Laminin B1 by cytoplasmic accumulation of La during epithelial to mesenchymal transition. Nucleic Acids Res 40(19):9738–9749. https://doi.org/10.1093/nar/gks760
Pozzi A, Yurchenco PD, Iozzo RV (2017) The nature and biology of basement membranes. Matrix Biol 57–58:1–11. https://doi.org/10.1016/j.matbio.2016.12.009
Rizvi S, Wang J, El-Khoueiry AB (2020) Liver cancer immunity. Hepatology 73(Suppl 1):86–103. https://doi.org/10.1002/hep.31416
Ruan H, Hao S, Young P, Zhang H (2015) Targeting cathepsin B for cancer therapies. Horiz Cancer Res 56:23–40
Sangro B, Chan SL, Meyer T, Reig M, El-Khoueiry A, Galle PR (2020) Diagnosis and management of toxicities of immune checkpoint inhibitors in hepatocellular carcinoma. J Hepatol 72(2):320–341. https://doi.org/10.1016/j.jhep.2019.10.021
Siegel RL, Miller KD, Jemal A (2020) Cancer statistics. CA Cancer J Clin 70(1):7–30. https://doi.org/10.3322/caac.21590
Sun Y, Zhang W, Bi X et al (2022) Systemic therapy for hepatocellular carcinoma: chinese consensus-based interdisciplinary expert statements. Liver Cancer 11(3):192–208. https://doi.org/10.1159/000521596
Tao J, Li X, Liang C, Liu Y, Zhou J (2022) Expression of basement membrane genes and their prognostic significance in clear cell renal cell carcinoma patients. Front Oncol 12:1026331. https://doi.org/10.3389/fonc.2022.1026331
Tátrai P, Dudás J, Batmunkh E et al (2006) Agrin, a novel basement membrane component in human and rat liver, accumulates in cirrhosis and hepatocellular carcinoma. Lab Investig 86(11):1149–1160. https://doi.org/10.1038/labinvest.3700475
Wan RJ, Peng W, Xia QX, Zhou HH, Mao XY (2021) Ferroptosis-related gene signature predicts prognosis and immunotherapy in glioma. CNS Neurosci Ther 27(8):973–986. https://doi.org/10.1111/cns.13654
Wang Q, Yu T, Yuan Y et al (2013) Sorafenib reduces hepatic infiltrated regulatory T cells in hepatocellular carcinoma patients by suppressing TGF-beta signal. J Surg Oncol 107(4):422–427. https://doi.org/10.1002/jso.23227
Xu Z, Peng B, Liang Q et al (2021) Construction of a ferroptosis-related nine-lncRNA signature for predicting prognosis and immune response in hepatocellular carcinoma. Front Immunol 12:719175. https://doi.org/10.3389/fimmu.2021.719175
Zhang B, Tang B, Gao J, Li J, Kong L, Qin L (2020) A hypoxia-related signature for clinically predicting diagnosis, prognosis and immune microenvironment of hepatocellular carcinoma patients. J Transl Med 18(1):342. https://doi.org/10.1186/s12967-020-02492-9
Zhang J, Yu S, Li Q, Wang Q, Zhang J (2022) Increased co-expression of MEST and BRCA1 is associated with worse prognosis and immune infiltration in ovarian cancer. Gynecol Oncol 164(3):566–576. https://doi.org/10.1016/j.ygyno.2022.01.010
Acknowledgements
This work was supported by the "Natural Science Foundation of Fujian Province, China" (No. 2020J011264), which provided funds for research to Hongbin Chen. The authors thank Dr. Yingming Sun for providing technical guidance.
Funding
This work was supported by the "Natural Science Foundation of Fujian Province, China" (No. 2020J011264), which provided funds for research to Hongbin Chen.
Author information
Authors and Affiliations
Contributions
HC and YZ in designed the study. YZ and ZY conducted the bioinformatics analysis. YZ drafted the manuscript. YC and KH drew the figures. ZY and YD prepared the tables. HC and YZ reviewed and revised the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The manuscript has been read and approved by all the authors.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

432_2022_4549_MOESM1_ESM.jpg
Figure S1: Construction of a BM-related genes risk signature. (A) LASSO coefficient profiles for the 22 DEGs. (B) LASSO deviance profiles. (JPG 650 kb)

432_2022_4549_MOESM2_ESM.jpg
Figure S2: Association between BM-related DEGs and immune. (A) Immune cells infiltration between high- and low-risk groups. (B) The relationships between the BMGs and immune infiltration cells. (JPG 5602 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, Y., Yin, Z., Huang, K. et al. The basement membrane-related gene signature is associated with immunity and predicts survival accurately in hepatocellular carcinoma. J Cancer Res Clin Oncol 149, 5751–5760 (2023). https://doi.org/10.1007/s00432-022-04549-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04549-2