Abstract
Objectives
More and more evidences show that circular RNAs (circRNAs) can be used as miRNA sponge to regulate the drug resistance of malignancies, including melanoma. However, how exosomal circRNAs participate in the therapeutic resistance of melanoma remains ambiguous.
Methods
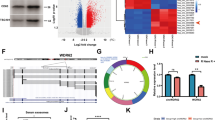
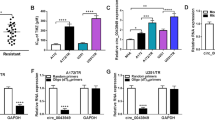
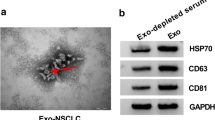
Vemurafenib-resistant A375 cells were cultured and then the circRNA profile of exosomes from the parental A375 and A375-resistant cells were sequenced. Transmission electron microscopy (TEM), exogenous nanoparticle tracking analysis (NTA) and Western Blot assays were leveraged to confirm the successful collection of exosomes from A375 and A375R cells. Another five published RNA-seq data and microRNA-seq data, and seven miRNA databases were collected to construct a competing endogenous RNA (ceRNA) network. Comprehensive bioinformatic analysis was adopted to identify key molecules related to the drug resistance, including multiscale embedded gene co-expression network analysis (MEGENA). Then, qRT-PCR, cell viability and colony formation were used to estimate the function of hub circRNAs. The role of has_circ_0001005 in vivo was verified via xenograft assay. The Tumor online Prognostic analyses Platform (ToPP) was leveraged to develop the has_circ_0001005-related prognostic models for melanoma patients based on TCGA data.
Results
Compared with parental cells, hsa_circ_0001005 expression was significantly increased in resistant cells and their exosomes. The elevated level of hsa_circ_0001005 was related to the poor clinical prognosis of melanoma patients. Hsa_circ_0001005 found in melanoma was mainly secreted by drug-resistant cells as exosomes. Exosomal hsa_circ_0001005 activated multiple canonical pathways related to drug resistance through sponging four miRNAs, thus suppressing the drug sensitivity of melanoma. Knocking down hsa_circ_0001005 in vitro, we found that the inhibition of hsa_circ_0001005 could hinder the clone formation of melanoma. Further in vivo animal experiments suggested that suppression of hsa_circ_0001005 can increase the sensitivity to Vemurafenib of melanoma cells. Finally, we also constructed the functional regulatory ceRNA network and prognostic risk models for hsa_circ_0001005, and further survival analysis reveals that the regulatory network and prognostic risk models obviously affected the prognosis of melanoma patients.
Conclusions
This study confirmed that the level of hsa_circ_0001005 in exosomes is the key factor affecting drug resistance of melanoma, which provides a new potential therapeutic target for melanoma patients.







Similar content being viewed by others
Data availability
All data related to the research are included in the article or uploaded as supplementary information. The raw data used to support the findings of this study are available from the corresponding authors upon reasonable request.
Abbreviations
- GO:
-
Gene ontology
- MF:
-
Molecular function
- BP:
-
Biological process
- CC:
-
Cellular component
- MEGENA:
-
Multiscale embedded gene co-expression network analysis
- KEGG:
-
Kyoto encyclopedia of genes
- OS:
-
Overall survival
- RNA-seq:
-
RNA sequencing
- circRNA:
-
Circular RNA
- TCGA:
-
The Cancer Genome Atlas
- ToPP:
-
Tumor online Prognostic analyses Platform (http://www.biostatistics.online/topp/index.php)
References
Bandyopadhyay S, Mitra R (2009) TargetMiner: microRNA target prediction with systematic identification of tissue-specific negative examples. Bioinformatics 25(20):2625–2631
Brighton HE, Angus SP, Bo T et al (2018) New mechanisms of resistance to MEK inhibitors in melanoma revealed by intravital imaging. Cancer Res 78(2):542–557
Caporali S, Amaro A, Levati L et al (2019) miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J Exp Clin Cancer Res 38(1):272
Chen Y, Wang X (2020) miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res 48(D1):D127–D131
Chen G, Huang AC, Zhang W et al (2018) Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 560(7718):382–386
Chou CH, Shrestha S, Yang CD et al (2018) miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res 46(D1):D296–D302
Davis J, Wayman M (2022) Encorafenib and binimetinib combination therapy in metastatic melanoma. J Adv Pract Oncol 13(4):450–455
Du WW, Yang W, Liu E et al (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44(6):2846–2858
Felicetti F, Errico MC, Bottero L et al (2008) The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res 68(8):2745–2754
Felicetti F, De Feo A, Coscia C et al (2016) Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J Transl Med 14:56
Glazar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20(11):1666–1670
Gonzalez M, de Mayo Las Casas C, Oramas J et al (2021) Intermittent BRAF inhibition in advanced BRAF mutated melanoma results of a phase II randomized trial. Nat Commun 12(1):7008–10
Hood JL, San RS, Wickline SA (2011) Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res 71(11):3792–3801
Huser L, Sachindra S, Granados K et al (2018) SOX2-mediated upregulation of CD24 promotes adaptive resistance toward targeted therapy in melanoma. Int J Cancer 143(12):3131–3142
Iero M, Valenti R, Huber V et al (2008) Tumour-released exosomes and their implications in cancer immunity. Cell Death Differ 15(1):80–88
Krzywinski M, Schein J, Birol I et al (2009) Circos: an information aesthetic for comparative genomics. Genome Res 19(9):1639–1645
Lewis BP, Shih IH, Jones-Rhoades MW et al (2003) Prediction of mammalian microRNA targets. Cell 115(7):787–798
Li JH, Liu S, Zhou H et al (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42(Database issue):92–97
Lima LG, Chammas R, Monteiro RQ et al (2009) Tumor-derived microvesicles modulate the establishment of metastatic melanoma in a phosphatidylserine-dependent manner. Cancer Lett 283(2):168–175
Liu CX, Chen LL (2022) Circular RNAs: characterization, cellular roles, and applications. Cell 185(12):2016–2034
Long GV, Grob JJ, Nathan P et al (2016) Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol 17(12):1743–1754
Luke JJ, Flaherty KT, Ribas A et al (2017) Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 14(8):463–482
Miranda KC, Huynh T, Tay Y et al (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126(6):1203–1217
O’Neill K, Liao WW, Patel A et al (2018) TEsmall identifies small RNAs associated with targeted inhibitor resistance in melanoma. Front Genet 9:461
Peinado H, Aleckovic M, Lavotshkin S et al (2012) Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 18(6):883–891
Robert C, Grob JJ, Stroyakovskiy D et al (2019) Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N Engl J Med 381(7):626–636
Shang Q, Du H, Wu X et al (2022) FMRP ligand circZNF609 destabilizes RAC1 mRNA to reduce metastasis in acral melanoma and cutaneous melanoma. J Exp Clin Cancer Res 41(1):170
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Singleton KR, Crawford L, Tsui E et al (2017) Melanoma therapeutic strategies that select against resistance by exploiting MYC-driven evolutionary convergence. Cell Rep 21(10):2796–2812
Song WM, Zhang B (2015) Multiscale embedded gene co-expression network analysis. PLoS Comput Biol 11(11):e1004574
Sticht C, De La Torre C, Parveen A et al (2018) miRWalk: an online resource for prediction of microRNA binding sites. PLoS One 13(10):e0206239
Su Y, Ko ME, Cheng H et al (2020) Multi-omic single-cell snapshots reveal multiple independent trajectories to drug tolerance in a melanoma cell line. Nat Commun 11(1):2345
Tan Y, Tang F, Li J et al (2022) Tumor-derived exosomes: the emerging orchestrators in melanoma. Biomed Pharmacother 149:112832
Thery C, Zitvogel L, Amigorena S (2002) Exosomes: composition, biogenesis and function. Nat Rev Immunol 2(8):569–579
Valadi H, Ekstrom K, Bossios A et al (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Vellano CP, White MG, Andrews MC et al (2022) Androgen receptor blockade promotes response to BRAF/MEK-targeted therapy. Nature 606(7915):797–803
Wang XT, Xie YB, Xiao Q (2011) Lentivirus-mediated RNA interference targeting E2F–1 inhibits human gastric cancer MGC-803 cell growth in vivo. Exp Mol Med 43(11):638–645
Wang TB, Geng M, Jin H et al (2021) SREBP1 site 1 protease inhibitor PF-429242 suppresses renal cell carcinoma cell growth. Cell Death Dis 12(8):717
Webber J, Steadman R, Mason MD et al (2010) Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res 70(23):9621–9630
Xia S, Feng J, Chen K et al (2018) CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res 46(D1):D925–D929
Xiao D, Barry S, Kmetz D et al (2016) Melanoma cell-derived exosomes promote epithelial-mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett 376(2):318–327
Yu G, Wang LG, Han Y et al (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287
Yu T, Liu K, Wu Y et al (2014) MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/beta-catenin signaling pathway. Oncogene 33(42):5017–5027
Zhang H, Deng T, Liu R et al (2017) Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun 8:15016
Zhou PH, Zheng JB, Wei GB et al (2015) Lentivirus-mediated RASSF1A expression suppresses aggressive phenotypes of gastric cancer cells in vitro and in vivo. Gene Ther 22(10):793–801
Funding
This study was funded by New Area Science and Technology Development Fund of Shanghai Pudong (No. PKJ2018-Y01).
Author information
Authors and Affiliations
Contributions
WX analyzed the data and wrote the manuscript. QC designed and directed this project, completed biological experiments, exosome chip sequencing, and participated in the revision and editing of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethical approval
All animal procedures are approved by Shanghai East Hospital and conform to the policy of Animal Ethics Committee of Shanghai East Hospital on the care, welfare and treatment of experimental animals. All participants consented to participate in the research before participating.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, X., Cheng, Q. Suppression of exosomal hsa_circ_0001005 eliminates the Vemurafenib resistance of melanoma. J Cancer Res Clin Oncol 149, 5921–5936 (2023). https://doi.org/10.1007/s00432-022-04434-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04434-y