Abstract
Background
High tumor mutation burden (TMB) failed to serve as a favorable prognostic biomarker for immunotherapy across all tumors. This study aimed to explore TMB-sensitive tumors on a pan-cancer level and construct their immune infiltration phenotypes in TMB-high groups.
Methods
Pan-cancer patients were separated into TMB-high and TMB-low groups based on the median TMB values per tumor. TMB-related genes were identified using differently expressed genes (DEGs) and differently mutated genes (DMGs) between the above two TMB groups. CIBERSORT algorithm was used to estimate the abundance of 22 tumor immune infiltrating cells (TIICs). Consensus clustering analysis was applied to predict molecular subtypes. Cox regression analysis was performed to evaluate the correlations between hub genes and TIICs and immunomodulator genes.
Results
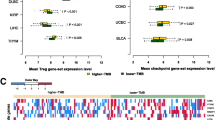
Nine TMB-sensitive tumors were identified by high-frequency of TMB-related genes. A total of 126 tumor-specific hub genes (1 in BLCA, 19 in BRCA, 4 in COAD, 4 in HNSC, 25 in LUAD, 2 in LUSC, 27 in SKCM, 37 in STAD, and 7 UCEC) were identified. In five out of nine TMB-sensitive tumors, the molecular subtypes based on hub gene expression were characterized by TMB values, prognostic values and tumor-specific TIICs levels. In TMB-high groups, hub genes associated immune infiltration phenotypes were constructed with key TIICs and immunomodulators spanning TMB-sensitive tumors.
Conclusions
Our tumor-specific analysis revealed hub genes associated immune infiltration features may serve as potential therapeutic targets and prognostic markers of immunotherapy, providing the potential underlying mechanism of immune infiltration in TMB-high groups across TMB-sensitive tumors.






Similar content being viewed by others
Data availability
Data are available in a public, open access repository. The data sets analyzed during the current study are available in The Cancer Genome Atlas (TCGA) (http://gdac.broadinstitute.org).
Abbreviations
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast invasive carcinoma
- COAD:
-
Colon adenocarcinoma
- ESCA:
-
Esophageal carcinoma
- HNSC:
-
Head and neck squamous cell carcinoma
- KIRC:
-
Kidney renal clear cell carcinoma
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PRAD:
-
Prostate adenocarcinoma
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- THYM:
-
Thymoma
- UCEC:
-
Uterine corpus endometrial carcinoma
References
Barroso-Sousa R, Jain E, Cohen O et al (2020a) Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol 31:387–394. https://doi.org/10.1016/j.annonc.2019.11.010
Barroso-Sousa R, Keenan TE, Pernas S et al (2020b) Tumor mutational burden and PTEN alterations as molecular correlates of response to PD-1/L1 blockade in metastatic triple-negative breast cancer. Clin Cancer Res 26:2565–2572. https://doi.org/10.1158/1078-0432.CCR-19-3507
Bouzidi L, Triki H, Charfi S et al (2021) Prognostic value of natural killer cells besides tumor-infiltrating lymphocytes in breast cancer tissues. Clin Breast Cancer 21:e738–e747. https://doi.org/10.1016/j.clbc.2021.02.003
Cao D, Xu H, Xu X et al (2019) High tumor mutation burden predicts better efficacy of immunotherapy: a pooled analysis of 103078 cancer patients. Oncoimmunology 8:e1629258. https://doi.org/10.1080/2162402X.2019.1629258
Chalmers ZR, Connelly CF, Fabrizio D et al (2017) Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9:34. https://doi.org/10.1186/s13073-017-0424-2
Chan TA, Yarchoan M, Jaffee E et al (2019) Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol 30:44–56. https://doi.org/10.1093/annonc/mdy495
Endris V, Buchhalter I, Allgäuer M et al (2019) Measurement of tumor mutational burden (TMB) in routine molecular diagnostics: in silico and real-life analysis of three larger gene panels. Int J Cancer 144:2303–2312. https://doi.org/10.1002/ijc.32002
Fabrizio DA, George TJ, Dunne RF et al (2018) Beyond microsatellite testing: assessment of tumor mutational burden identifies subsets of colorectal cancer who may respond to immune checkpoint inhibition. J Gastrointest Oncol 9:610–617. https://doi.org/10.21037/jgo.2018.05.06
Fan S, Gao X, Qin Q et al (2020) Association between tumor mutation burden and immune infiltration in ovarian cancer. Int Immunopharmacol 89:107126. https://doi.org/10.1016/j.intimp.2020.107126
Fumet J-D, Truntzer C, Yarchoan M, Ghiringhelli F (2020) Tumour mutational burden as a biomarker for immunotherapy: current data and emerging concepts. Eur J Cancer 131:40–50. https://doi.org/10.1016/j.ejca.2020.02.038
Galluzzi L, Chan TA, Kroemer G et al (2018) The hallmarks of successful anticancer immunotherapy. Sci Transl Med 10:eaat7807. https://doi.org/10.1126/scitranslmed.aat7807
Gao G, Wang Z, Qu X, Zhang Z (2020) Prognostic value of tumor-infiltrating lymphocytes in patients with triple-negative breast cancer: a systematic review and meta-analysis. BMC Cancer 20:179. https://doi.org/10.1186/s12885-020-6668-z
Goodman AM, Kato S, Bazhenova L et al (2017) Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther 16:2598–2608. https://doi.org/10.1158/1535-7163.MCT-17-0386
Hashemi S, Fransen MF, Niemeijer A et al (2021) Surprising impact of stromal TIL’s on immunotherapy efficacy in a real-world lung cancer study. Lung Cancer 153:81–89. https://doi.org/10.1016/j.lungcan.2021.01.013
Hellmann MD, Callahan MK, Awad MM et al (2019) Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 35:329. https://doi.org/10.1016/j.ccell.2019.01.011
Hellmann MD, Ciuleanu T-E, Pluzanski A et al (2018a) Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med 378:2093–2104. https://doi.org/10.1056/NEJMoa1801946
Hellmann MD, Nathanson T, Rizvi H et al (2018b) Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33:843-852.e4. https://doi.org/10.1016/j.ccell.2018.03.018
Hu Q, Hong Y, Qi P et al (2021) Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat Commun 12:2186. https://doi.org/10.1038/s41467-021-22300-2
Khasraw M, Walsh KM, Heimberger AB, Ashley DM (2020) What is the burden of proof for tumor mutational burden in gliomas? Neuro Oncol. https://doi.org/10.1093/neuonc/noaa256
Kim J, Kim B, Kang SY et al (2020) Tumor mutational burden determined by panel sequencing predicts survival after immunotherapy in patients with advanced gastric cancer. Front Oncol 10:314. https://doi.org/10.3389/fonc.2020.00314
Klempner SJ, Fabrizio D, Bane S et al (2020) Tumor mutational burden as a predictive biomarker for response to immune checkpoint inhibitors: a review of current evidence. Oncologist 25:e147–e159. https://doi.org/10.1634/theoncologist.2019-0244
Le DT, Uram JN, Wang H et al (2015) PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 372:2509–2520. https://doi.org/10.1056/NEJMoa1500596
Lee C-H, Yelensky R, Jooss K, Chan TA (2018) Update on tumor neoantigens and their utility: why it is good to be different. Trends Immunol 39:536–548. https://doi.org/10.1016/j.it.2018.04.005
Li L, Bai L, Lin H et al (2021) Multiomics analysis of tumor mutational burden across cancer types. Comput Struct Biotechnol J 19:5637–5646. https://doi.org/10.1016/j.csbj.2021.10.013
Lin J, Lin Y, Huang Z, Li X (2020) Identification of prognostic biomarkers of cutaneous melanoma based on analysis of tumor mutation burden. Comput Math Methods Med 2020:8836493. https://doi.org/10.1155/2020/8836493
Marabelle A, Le DT, Ascierto PA et al (2020) Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 38:1–10. https://doi.org/10.1200/JCO.19.02105
Marcus L, Fashoyin-Aje LA, Donoghue M et al (2021) FDA approval summary: pembrolizumab for the treatment of tumor mutational burden-high solid tumors. Clin Cancer Res 27:4685–4689. https://doi.org/10.1158/1078-0432.CCR-21-0327
Mayakonda A, Lin D-C, Assenov Y et al (2018) Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 28:1747–1756. https://doi.org/10.1101/gr.239244.118
McGrail DJ, Pilié PG, Rashid NU et al (2021) High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 32:661–672. https://doi.org/10.1016/j.annonc.2021.02.006
Mielgo-Rubio X, Uribelarrea EA, Cortés LQ, Moyano MS (2021) Immunotherapy in non-small cell lung cancer: update and new insights. J Clin Transl Res 7:1–21
Mounir M, Lucchetta M, Silva TC et al (2019) New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput Biol 15:e1006701. https://doi.org/10.1371/journal.pcbi.1006701
Mouw KW, Goldberg MS, Konstantinopoulos PA, D’Andrea AD (2017) DNA damage and repair biomarkers of immunotherapy response. Cancer Discov 7:675–693. https://doi.org/10.1158/2159-8290.CD-17-0226
Nagahashi M, Shimada Y, Ichikawa H et al (2019) Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci 110:6–15. https://doi.org/10.1111/cas.13837
Nan Z, Guoqing W, Xiaoxu Y et al (2021) The predictive efficacy of tumor mutation burden (TMB) on nonsmall cell lung cancer treated by immune checkpoint inhibitors: a systematic review and meta-analysis. Biomed Res Int 2021:1780860. https://doi.org/10.1155/2021/1780860
Newman AM, Liu CL, Green MR et al (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457. https://doi.org/10.1038/nmeth.3337
Ritchie ME, Phipson B, Wu D et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Rizvi NA, Hellmann MD, Snyder A et al (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124–128. https://doi.org/10.1126/science.aaa1348
Samstein RM, Lee C-H, Shoushtari AN et al (2019) Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet 51:202–206. https://doi.org/10.1038/s41588-018-0312-8
Schrock AB, Ouyang C, Sandhu J et al (2019) Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol 30:1096–1103. https://doi.org/10.1093/annonc/mdz134
Shao C, Li G, Huang L et al (2020) Prevalence of high tumor mutational burden and association with survival in patients with less common solid tumors. JAMA Netw Open 3:e2025109. https://doi.org/10.1001/jamanetworkopen.2020.25109
Snyder A, Makarov V, Merghoub T et al (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199. https://doi.org/10.1056/NEJMoa1406498
Spector ME, Bellile E, Amlani L et al (2019) Prognostic value of tumor-infiltrating lymphocytes in head and neck squamous cell carcinoma. JAMA Otolaryngol Head Neck Surg 145:1012–1019. https://doi.org/10.1001/jamaoto.2019.2427
Subbiah V, Solit DB, Chan TA, Kurzrock R (2020) The FDA approval of pembrolizumab for adult and pediatric patients with tumor mutational burden (TMB) ≥10: a decision centered on empowering patients and their physicians. Ann Oncol 31:1115–1118. https://doi.org/10.1016/j.annonc.2020.07.002
Tamminga M, Hiltermann TJN, Schuuring E et al (2020) Immune microenvironment composition in non-small cell lung cancer and its association with survival. Clin Transl Immunology 9:e1142. https://doi.org/10.1002/cti2.1142
Tan S, Li D, Zhu X (2020) Cancer immunotherapy: pros, cons and beyond. Biomed Pharmacother 124:109821. https://doi.org/10.1016/j.biopha.2020.109821
Van Allen EM, Miao D, Schilling B et al (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350:207–211. https://doi.org/10.1126/science.aad0095
Vihervuori H, Autere TA, Repo H et al (2019) Tumor-infiltrating lymphocytes and CD8+ T cells predict survival of triple-negative breast cancer. J Cancer Res Clin Oncol 145:3105–3114. https://doi.org/10.1007/s00432-019-03036-5
Wang F, Wei XL, Wang FH et al (2019) Safety, efficacy and tumor mutational burden as a biomarker of overall survival benefit in chemo-refractory gastric cancer treated with toripalimab, a PD-1 antibody in phase Ib/II clinical trial NCT02915432. Ann Oncol 30:1479–1486. https://doi.org/10.1093/annonc/mdz197
Wang L, Yao Y, Xu C et al (2021) Exploration of the tumor mutational burden as a prognostic biomarker and related hub gene identification in prostate cancer. Technol Cancer Res Treat 20:15330338211052154. https://doi.org/10.1177/15330338211052154
Wang X, Li M (2019) Correlate tumor mutation burden with immune signatures in human cancers. BMC Immunol 20:4. https://doi.org/10.1186/s12865-018-0285-5
Wilkerson MD, Hayes DN (2010) ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 26:1572–1573. https://doi.org/10.1093/bioinformatics/btq170
Yarchoan M, Johnson BA, Lutz ER et al (2017) Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer 17:209–222. https://doi.org/10.1038/nrc.2016.154
Yin W, Jiang X, Tan J et al (2020) Development and validation of a tumor mutation burden-related immune prognostic model for lower-grade glioma. Front Oncol 10:1409. https://doi.org/10.3389/fonc.2020.01409
Zhang C, Li Z, Qi F, et al (2019a) Exploration of the relationships between tumor mutation burden with immune infiltrates in clear cell renal cell carcinoma. Ann Transl Med 7:648. https://doi.org/10.21037/atm.2019a.10.84
Zhang D, He W, Wu C et al (2019b) Scoring system for tumor-infiltrating lymphocytes and its prognostic value for gastric cancer. Front Immunol 10:71. https://doi.org/10.3389/fimmu.2019.00071
Zhao L, Fu X, Han X et al (2021) Tumor mutation burden in connection with immune-related survival in uterine corpus endometrial carcinoma. Cancer Cell Int 21:80. https://doi.org/10.1186/s12935-021-01774-6
Acknowledgements
We sincerely acknowledge the contributions from the TCGA project.
Funding
This work was supported by the National Natural Science Foundation of China (82073284); Shenzhen Key Medical Discipline Construction Fund (SZXK053); Guangdong Basic and Applied Basic Research Foundation (2021A1515012163).
Author information
Authors and Affiliations
Contributions
HuW and YC conceptualized and designed the study. All authors wrote the manuscript; HuW and HaW collected and analyzed the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Patient consent for publication
Not required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wu, H., Wang, H. & Chen, Y. Pan-cancer analysis of tumor mutation burden sensitive tumors reveals tumor-specific subtypes and hub genes related to immune infiltration. J Cancer Res Clin Oncol 149, 2793–2804 (2023). https://doi.org/10.1007/s00432-022-04139-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04139-2