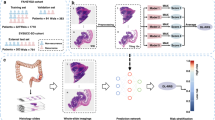
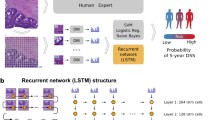
Abstract
Purpose
Most of Stage II/III colorectal cancer (CRC) patients can be cured by surgery alone, and only certain CRC patients benefit from adjuvant chemotherapy. Risk stratification based on deep-learning from haematoxylin and eosin (H&E) images has been postulated as a potential predictive biomarker for benefit from adjuvant chemotherapy. However, very limited success has been achieved in using biomarkers, including deep-learning-based markers, to facilitate the decision for adjuvant chemotherapy despite recent advances of artificial intelligence.
Methods
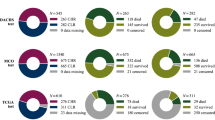
We trained and internally validated CRCNet using 780 Stage II/III CRC patients from Molecular and Cellular Oncology. Independent external validation of the model was performed using 337 Stage II/III CRC patients from The Cancer Genome Atlas (TCGA).
Results
CRCNet stratified the patients into high, medium, and low-risk subgroups. Multivariate Cox regression analyses confirmed that CRCNet risk groups are statistically significant after adjusting for existing risk factors. The high-risk subgroup significantly benefits from adjuvant chemotherapy. A hazard ratio (chemo-treated vs untreated) of 0.2 (95% Confidence Interval (CI), 0.05–0.65; P = 0.009) and 0.6 (95% CI 0.42–0.98; P = 0.038) are observed in the TCGA and MCO Fluorouracil-treated patients, respectively. Conversely, no significant benefit from chemotherapy is observed in the low- and medium-risk groups (P = 0.2–1).
Conclusion
The retrospective analysis provides further evidence that H&E image-based biomarkers may potentially be of great use in delivering treatments following surgery for Stage II/III CRC, improving patient survival, and avoiding unnecessary treatment and associated toxicity, and warrants further validation on other datasets and prospective confirmation in clinical trials.



Similar content being viewed by others
Data availability
The TCGA dataset is publicly available at the TCGA portal (https://portal.gdc.cancer.gov). The public TCGA clinical data is available at the website(https://xenabrowser.net/datapages/). Xception model weights are available at (https://github.com/fchollet/deep-learning-models/releases/download/v0.4/xception_weights_tf_dim_ordering_tf_kernels_notop.h5). The MCO dataset is available through the SREDH Consortium (www.sredhconsortium.org), which was used with permission in our current study.
Code availability
Source code is available at https://github.com/1996lixingyu1996/CRCNet.
References
A Package for Survival Analysis in R, 2020
André T, Boni C, Navarro M et al (2009) Improved overall survival with oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment in stage II or III colon cancer in the MOSAIC trial. J Clin Oncol 27(19):3109–3116. https://doi.org/10.1200/jco.2008.20.6771
Anonymous (2013) Data from: MCO study tumour collection. https://doi.org/10.4225/53/5559205bea135
Benson AB III, Venook AP, Cederquist L et al (2017) Colon Cancer, Version 1.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Comprehensive Cancer Netw JNCCN. 15(3):370–398. https://doi.org/10.6004/jnccn.2017.0036
Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A (2019) Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 16(11):703–715. https://doi.org/10.1038/s41571-019-0252-y
Compton CC, Greene FL (2004) The staging of colorectal cancer: 2004 and beyond. CA Cancer J Clin 54(6):295–308. https://doi.org/10.3322/canjclin.54.6.295
Danielsen HE, Hveem TS, Domingo E et al (2018) Prognostic markers for colorectal cancer: estimating ploidy and stroma. Ann Oncol 29(3):616–623. https://doi.org/10.1093/annonc/mdx794
de Gramont A, Van Cutsem E, Schmoll HJ et al (2012) Bevacizumab plus oxaliplatin-based chemotherapy as adjuvant treatment for colon cancer (AVANT): a phase 3 randomised controlled trial. Lancet Oncol 13(12):1225–1233. https://doi.org/10.1016/s1470-2045(12)70509-0
Galon J, Mlecnik B, Bindea G et al (2014) Towards the introduction of the “Immunoscore” in the classification of malignant tumours. J Pathol 232(2):199–209. https://doi.org/10.1002/path.4287
Gray RG, Quirke P, Handley K et al (2011) Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 29(35):4611–4619. https://doi.org/10.1200/jco.2010.32.8732
Guinney J, Dienstmann R, Wang X et al (2015) The consensus molecular subtypes of colorectal cancer. Nat Med 21(11):1350–1356. https://doi.org/10.1038/nm.3967
Jemal A, Tiwari RC, Murray T et al (2004) Cancer statistics, 2004. CA Cancer J Clin 54(1):8–29. https://doi.org/10.3322/canjclin.54.1.8
Jonnagaddala J, Croucher JL, Jue TR et al (2016) Integration and analysis of heterogeneous colorectal cancer data for translational research. Nurs Inf 225:387–391
Kather JN, Krisam J, Charoentong P et al (2019) Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med 16(1):e1002730. https://doi.org/10.1371/journal.pmed.1002730
Mlecnik B, Bifulco C, Bindea G et al (2020) Multicenter international society for immunotherapy of cancer study of the consensus immunoscore for the prediction of survival and response to chemotherapy in stage III colon cancer. J Clin Oncol 38(31):3638–3651. https://doi.org/10.1200/jco.19.03205
Muzny DM, Bainbridge MN, Chang K et al (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature 487(7407):330–337. https://doi.org/10.1038/nature11252
O’Connell MJ, Lavery I, Yothers G et al (2010) Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol 28(25):3937–3944. https://doi.org/10.1200/jco.2010.28.9538
Reinert T, Henriksen TV, Christensen E et al (2019) Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol 5(8):1124–1131. https://doi.org/10.1001/jamaoncol.2019.0528
Schmoll HJ, Van Cutsem E, Stein A et al (2012) ESMO Consensus Guidelines for management of patients with colon and rectal cancer. A personalized approach to clinical decision making. Ann Oncol 23(10):2479–2516. https://doi.org/10.1093/annonc/mds236
Schmoll HJ, Tabernero J, Maroun J et al (2015) Capecitabine plus oxaliplatin compared with fluorouracil/folinic acid as adjuvant therapy for stage III colon cancer: final results of the NO16968 randomized controlled phase III trial. J Clin Oncol 33(32):3733–3740. https://doi.org/10.1200/jco.2015.60.9107
Schrag D, Rifas-Shiman S, Saltz L, Bach PB, Begg CB (2002) Adjuvant chemotherapy use for Medicare beneficiaries with stage II colon cancer. J Clin Oncol 20(19):3999–4005. https://doi.org/10.1200/jco.2002.11.084
Skrede OJ, De Raedt S, Kleppe A et al (2020) Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet (london, England) 395(10221):350–360. https://doi.org/10.1016/s0140-6736(19)32998-8
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Tie J, Cohen JD, Wang Y et al (2019) Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol 5(12):1710–1717. https://doi.org/10.1001/jamaoncol.2019.3616
Weiss JM, Schumacher J, Allen GO et al (2014) Adjuvant chemotherapy for stage II right-sided and left-sided colon cancer: analysis of SEER-medicare data. Ann Surg Oncol 21(6):1781–1791. https://doi.org/10.1245/s10434-014-3631-8
Wulczyn E, Steiner DF, Moran M et al (2021) Interpretable survival prediction for colorectal cancer using deep learning. Npj Digit Med 4(1):71. https://doi.org/10.1038/s41746-021-00427-2
Yang L, Chen P, Zhang L et al (2020) Prognostic value of nucleotyping, DNA ploidy and stroma in high-risk stage II colon cancer. Br J Cancer 123(6):973–981. https://doi.org/10.1038/s41416-020-0974-8
Yao J, Zhu X, Jonnagaddala J, Hawkins N, Huang J (2020) Whole slide images based cancer survival prediction using attention guided deep multiple instance learning networks. Med Image Anal 65:101789. https://doi.org/10.1016/j.media.2020.101789
Yothers G, O’Connell MJ, Allegra CJ et al (2011) Oxaliplatin as adjuvant therapy for colon cancer: updated results of NSABP C-07 trial, including survival and subset analyses. J Clin Oncol 29(28):3768–3774. https://doi.org/10.1200/jco.2011.36.4539
Yothers G, O’Connell MJ, Lee M et al (2013) Validation of the 12-gene colon cancer recurrence score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol 31(36):4512–4519. https://doi.org/10.1200/jco.2012.47.3116
Acknowledgements
The research of Xingyu Li, Shuhua Yang, and Hong Zhang was partially supported by National Natural Science Foundation of China (No. 11771096, 72091212), Anhui Center for Applied Mathematics, and Special Project of Strategic Leading Science and Technology of CAS (No. XDC08010100). Jitendra Jonnagaddala is funded by the Australian National Health and Medical Research Council (No. GNT1192469). Jitendra Jonnagaddala is also supported by the Google Cloud Research Credits program( No. GCP19980904). We thank Michelle Xu (Princeton Day School) for writing assistance and language editing.
Funding
National Natural Science Foundation of China (No. 11771096, 72091212, 12171451), Anhui Center for Applied Mathematics, and Special Project of Strategic Leading Science and Technology of CAS (No. XDC08010100). Australian National Health and Medical Research Council (No. GNT1192469). Google Cloud Research Credits program with the award (No. GCP19980904).
Author information
Authors and Affiliations
Contributions
XSX, XL, and HZ contributed to design of the research; JJ, XL, and XSX contributed to data acquisition; XL, XSX, and SY contributed to data analysis. XL, XSX, JJ, and HZ contributed to data interpretation. XL, XSX, JJ, and HZ wrote the manuscript; and all authors critically reviewed the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Software
All statistical analysis were conducted in R (version 4.1.0) unless otherwise specified. The following libraries were used: survminer (version 0.4.9), survival (version 3.2–13), and ggplot2 (version 3.3.5). The U-Net model, tissue classifier, risk group predictor were trained with Python (version 3.7.9), Tf-nightly-gpu (version 2.5.0.dev20210209), scipy (version 1.6.1), scikit-learn (version 0.24.1), openslide-python (version 1.1.2), opencv-python (version 4.5.1.48), numpy (version 1.19.5), numba (version 0.52.0), matplotlib (version 3.3.4), pandas (version 1.2.2), and torchvision (version 0.8.2). All training parameters were provided in the source code available at https://github.com/1996lixingyu1996/CRCNet. Source code is available at https://github.com/1996lixingyu1996/CRCNet.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, X., Jonnagaddala, J., Yang, S. et al. A retrospective analysis using deep-learning models for prediction of survival outcome and benefit of adjuvant chemotherapy in stage II/III colorectal cancer. J Cancer Res Clin Oncol 148, 1955–1963 (2022). https://doi.org/10.1007/s00432-022-03976-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-03976-5