Abstract
Introduction
Increasing evidence indicates an important role of microbiota in cancer development and progression, while little is known about the correlation between microbiota and renal cell carcinoma (RCC). Thus, we performed this study to profile the intratumoral microbiota possibly associated with RCC.
Materials and methods
Paired RCC and adjacent normal tissue samples were collected from 24 patients with RCC. V3-V4 variable region of microbial 16S rRNA gene was sequenced using Illumina MiSeq. Sequencing reads were processed using QIIME. Differentially abundant bacterial taxa between groups were identified by LEfSe, and their potential functions were inferred by PICRUSt.
Results
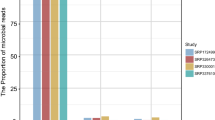
Decreased species diversity was presented in RCC tissues (Simpson index, P = 0.0340), and the composition of the bacterial community in RCC tissues was significantly distinct from that in normal tissues (unweighted UniFrac distance, P = 0.026; weighted UniFrac distance, P = 0.017). Compared with normal tissues, 25 taxa increased and 47 reduced taxa were identified in RCC tissues. Among these taxa, the class Chloroplast (AUC = 0.91, P < 0.0001) and the order Streptophyta (AUC = 0.89, P < 0.0001) showed high indication accuracy to discriminate RCC tissues from normal tissues. Furthermore, nine altered pathways were identified in RCC tissues to reveal the potential microbial function.
Conclusions
Our results have uncovered the presence of distinct microbiota in RCC and adjacent normal tissues and provided a better understanding of the possible role of the intratumoral microbiota in RCC. Further studies are required to confirm our results and determine the real correlation between microbiota and RCC.





Similar content being viewed by others
References
Ahn J et al (2013) Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105:1907–1911. https://doi.org/10.1093/jnci/djt300
Apopa PL et al (2018) PARP1 Is up-regulated in non-small cell lung cancer tissues in the presence of the cyanobacterial toxin microcystin. Front Microbiol 9:1757. https://doi.org/10.3389/fmicb.2018.01757
Chen H, Jiang W (2014) Application of high-throughput sequencing in understanding human oral microbiome related with health and disease. Front Microbiol 5:508. https://doi.org/10.3389/fmicb.2014.00508
Chen W, Liu F, Ling Z, Tong X, Xiang C (2012) Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PloS one 7:e39743. https://doi.org/10.1371/journal.pone.0039743
Cogdill AP, Gaudreau PO, Arora R, Gopalakrishnan V, Wargo JA (2018) The impact of intratumoral and gastrointestinal microbiota on systemic cancer therapy. Trends Immunol 39:900–920. https://doi.org/10.1016/j.it.2018.09.007
Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayrede JP (2010) Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc Natl Acad Sci USA 107:11537–11542. https://doi.org/10.1073/pnas.1001261107
Derosa L et al (2020) Gut bacteria composition drives primary resistance to cancer immunotherapy in renal cell carcinoma patients. Eur Urol 78:195–206. https://doi.org/10.1016/j.eururo.2020.04.044
Dizman N, Philip EJ, Pal SK (2020) Genomic profiling in renal cell carcinoma Nature reviews. Nephrology 16:435–451. https://doi.org/10.1038/s41581-020-0301-x
Eckburg PB et al (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638. https://doi.org/10.1126/science.1110591
Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR (2010) The human nasal microbiota and Staphylococcus aureus carriage. PloS One 5:e10598. https://doi.org/10.1371/journal.pone.0010598
Gill SR et al (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359. https://doi.org/10.1126/science.1124234
Golebiewski M, Tretyn A (2020) Generating amplicon reads for microbial community assessment with next-generation sequencing. J Appl Microbiol 128:330–354. https://doi.org/10.1111/jam.14380
Grice EA, Segre JA (2011) The skin microbiome. Nature Rev Microbiol 9:244–253. https://doi.org/10.1038/nrmicro2537
Huang JJ, Hsieh JJ (2020) The therapeutic landscape of renal cell carcinoma: from the dark age to the golden age. . Seminars Nephrol 40:28–41. https://doi.org/10.1016/j.semnephrol.2019.12.004
Khanna S, Tosh PK (2014) A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin Proceed 89:107–114. https://doi.org/10.1016/j.mayocp.2013.10.011
Kim TK et al (2009) Heterogeneity of vaginal microbial communities within individuals. J Clin Microbiol 47:1181–1189. https://doi.org/10.1128/JCM.00854-08
Knauf F, Brewer JR, Flavell RA (2019) Immunity, microbiota and kidney disease. Nature Rev Nephrol 15:263–274. https://doi.org/10.1038/s41581-019-0118-7
Kotecha RR, Motzer RJ, Voss MH (2019) Towards individualized therapy for metastatic renal cell carcinoma Nature reviews. Clin Oncol 16:621–633. https://doi.org/10.1038/s41571-019-0209-1
Lalani AA et al (2020) Effect of antibiotic use on outcomes with systemic therapies in metastatic renal cell carcinoma. Eur Urol Oncol 3:372–381. https://doi.org/10.1016/j.euo.2019.09.001
Langille MG et al (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnol 31:814–821. https://doi.org/10.1038/nbt.2676
Liss MA et al (2020) Microbiome within primary tumor tissue from renal cell carcinoma may be associated with PD-L1 expression of the venous tumor thrombus. Adv Urol. https://doi.org/10.1155/2020/9068068
McQuade JL, Daniel CR, Helmink BA, Wargo JA (2019) Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol 20:e77–e91. https://doi.org/10.1016/S1470-2045(18)30952-5
Mei QX, Huang CL, Luo SZ, Zhang XM, Zeng Y, Lu YY (2018) Characterization of the duodenal bacterial microbiota in patients with pancreatic head cancer vs. healthy controls. Pancreatology 18:438–445. https://doi.org/10.1016/j.pan.2018.03.005
Pljesa-Ercegovac M, Savic-Radojevic A, Coric V, Radic T, Simic T (2020) Glutathione transferase genotypes may serve as determinants of risk and prognosis in renal cell carcinoma. BioFactors 46:229–238. https://doi.org/10.1002/biof.1560
Raja R, Hemaiswarya S, Ganesan V, Carvalho IS (2016) Recent developments in therapeutic applications of Cyanobacteria. Crit Rev Microbiol 42:394–405. https://doi.org/10.3109/1040841X.2014.957640
Shin NR, Whon TW, Bae JW (2015) Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 33:496–503. https://doi.org/10.1016/j.tibtech.2015.06.011
Siegel RL, Miller KD (2019) Jemal A (2019) Cancer statistics. CA Cancer J Clin 69:7–34. https://doi.org/10.3322/caac.21551
Thorell K et al (2017) In vivo analysis of the viable microbiota and helicobacter pylori transcriptome in gastric infection and early stages of carcinogenesis. Infec Immunity. https://doi.org/10.1128/IAI.00031-17
Tucker MD, Rini BI (2020) Predicting response to immunotherapy in metastatic renal cell carcinoma. Cancers. https://doi.org/10.3390/cancers12092662
Ueda K et al (2019) The impact of antibiotics on prognosis of metastatic renal cell carcinoma in Japanese patients treated with immune checkpoint inhibitors. Anticancer Res 39:6265–6271. https://doi.org/10.21873/anticanres.13836
Usher-Smith J, Simmons RK, Rossi SH, Stewart GD (2020) Current evidence on screening for renal cancer Nature reviews. Urology. https://doi.org/10.1038/s41585-020-0363-3
Wu S et al (2009) A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nature Med 15:1016–1022. https://doi.org/10.1038/nm.2015
Wu P et al (2018) Profiling the urinary microbiota in male patients with bladder cancer in China. . Front Cell Infec Microbiol 8:167. https://doi.org/10.3389/fcimb.2018.00167
Funding
This research was supported by the National Natural Science Foundation of China (Grant No. 32000650). Henan Provincial Scientific and Technological Research Project (Grant No. 192102310036), Henan Provincial Medical Scientific and Technological Research Project (Grant No. 201702191). The sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Author information
Authors and Affiliations
Contributions
Junpeng Wang, Xin Li, Kangdong Liu, and Tianzhong Yan conceived and designed the study. Junpeng Wang, Xin Li, Xiaoqiang Wu, and Zhiwei Wang performed the study and collected data. Junpeng Wang, Xin Li, Chan Zhang, and Guanghui Cao performed the analysis. Junpeng Wang, and Xin Li drafted, edited and revised the manuscript. Kangdong Liu and Tianzhong Yan supervised the study.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Li, X., Wu, X. et al. Uncovering the microbiota in renal cell carcinoma tissue using 16S rRNA gene sequencing. J Cancer Res Clin Oncol 147, 481–491 (2021). https://doi.org/10.1007/s00432-020-03462-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-020-03462-w