Abstract
Purpose
Studies on the performance of epigenetic-based biomarkers in colorectal cancer (CRC) are scarce and have shown contradictory results. Thus, we sought to examine the prognostic value of histone-modifying enzymes (EZH2, SETDB1 and LSD-1) and histone post-translational marks (H3K27me3 and H3K9me3) in CRC.
Methods
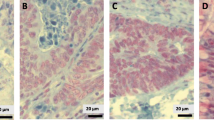
A retrospective series of 207 CRC patients primarily submitted to surgery in a cancer center was included in this study. Clinicopathological data were retrieved. One representative paraffin block per case was selected for immunohistochemistry, including normal and CRC tissues whenever possible. The percentage of positive nuclear staining (digital image analysis) was used to classify patients into “low” and “high” expression groups for each biomarker. Correlations between immunoexpression levels, clinicopathological features and clinical outcomes [disease-specific (DSS) and disease-free (DFS) survival] were examined. Statistical significance was set at p < 0.05.
Results
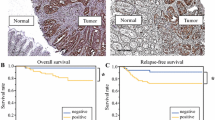
CRC tissues showed significantly lower expression of SETDB1 and higher expression of the remainder four biomarkers compared to normal mucosa. High EZH2 expression correlated with disease recurrence/progression, whereas low LSD1 expression and high H3K9me3 and H3K27me3 expression were associated with more advanced stage. In multivariable analysis, cases with high LSD1 expression displayed significantly better DSS and DFS (HR 0.477, 95% confidence interval: 0.247–0.923) adjusted for pathological TNM stage.
Conclusion
EZH2, SETDB1, LSD1, H3K9me3 and H3K27me3 expression are altered in CRC and may play a role in colorectal carcinogenesis. LSD1 immunoexpression levels independently predicted patient outcome in this cohort. Further investigations, using larger series, are warranted to confirm its potential clinical value and unravel underlying molecular mechanisms.



Similar content being viewed by others
Abbreviations
- CI:
-
Confidence interval
- CRC:
-
Colorectal cancer
- DFS:
-
Disease-free survival
- DSS:
-
Disease-specific survival
- EZH2:
-
Enhancer of zeste homolog 2
- HR:
-
Hazard ratio
- H3K9me3:
-
Histone H3 lysine 9 trimethylation
- H3K27me3:
-
Histone H3 lysine 27 trimethylation
- IHC:
-
Immunohistochemistry
- LSD-1:
-
Lysine-specific demethylase-1
- NOS:
-
Not otherwise specified
- SETDB1:
-
SET domain bifurcated 1
References
Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F (2017) Global patterns and trends in colorectal cancer incidence and mortality. Gut 66:683–691. https://doi.org/10.1136/gutjnl-2015-310912
Benard A, Goossens-Beumer IJ, van Hoesel AQ, de Graaf W, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ (2014a) Histone trimethylation at H3K4, H3K9 and H4K20 correlates with patient survival and tumor recurrence in early-stage colon cancer. BMC Cancer 14:531. https://doi.org/10.1186/1471-2407-14-531
Benard A, Goossens-Beumer IJ, van Hoesel AQ, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ (2014b) Prognostic Value of Polycomb Proteins EZH2, BMI1 and SUZ12 and Histone Modification H3K27me3 in Colorectal Cancer. PLoS One 9:e108265. https://doi.org/10.1371/journal.pone.0108265
Chen J, Ding J, Wang Z, Zhu J, Wang X, Du J (2017) Identification of downstream metastasis-associated target genes regulated by LSD1 in colon cancer cells. Oncotarget 8:19609–19630. https://doi.org/10.18632/oncotarget.14778
Deb G, Singh AK, Gupta S (2014) EZH2: not EZHY (easy) to deal. Mol Cancer Res 12:639–653. https://doi.org/10.1158/1541-7786.MCR-13-0546
Derks S, Bosch LJ, Niessen HE, Moerkerk PT, van der Bosch SM, Carvalho B, Mongera S, Voncken JW, Meijer GA, de Bruine AP, Herman JG, van Engeland M (2009) Promoter CpG island hypermethylation-and H3K9me3 and H3K27me3-mediated epigenetic silencing targets the deleted in colon cancer (DCC) gene in colorectal carcinogenesis without affecting neighboring genes on chromosomal region 18q21. Carcinogenesis 30:1041–1048. https://doi.org/10.1093/carcin/bgp073
Ding J, Zhang ZM, Xia Y, Liao GQ, Pan Y, Liu S, Zhang Y, Yan ZS (2013) LSD1-mediated epigenetic modification contributes to proliferation and metastasis of colon cancer. Br J Cancer 109:994–1003. https://doi.org/10.1038/bjc.2013.364
Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F (2015) Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 136:E359–E386. https://doi.org/10.1002/ijc.29210
Flavahan WA, Gaskell E, Bernstein BE (2017) Epigenetic plasticity and the hallmarks of cancer. Science 357:eaal2380. https://doi.org/10.1126/science.aal2380
Fluge Ø, Gravdal K, Carlsen E, Vonen B, Kjellevold K, Refsum S, Lilleng R, Eide TJ, Halvorsen TB, Tveit KM, Otte AP, Akslen LA, Dahl O, Norwegian Gastrointestinal Cancer Group (2009) Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br J Cancer 101:1282–1289. https://doi.org/10.1038/sj.bjc.6605333
Hamilton SR, Bosman FT, Boffetta P, Ilyas M, Morreau H, Nakamura SI, Quirke P, Riboli E, Sobin LH (2010) Carcinoma of the colon and rectum. In: Bosman FT, Carneiro F, Hruban RH, Theise ND (eds) WHO Classification of tumours of the digestive system, 4th edn. IARC, Lyon 2010, pp 134–146
Hino S, Kohrogi K, Nakao M (2016) Histone demethylase LSD1 controls the phenotypic plasticity of cancer cells. Cancer Sci 107:1187–1192. https://doi.org/10.1111/cas.13004
Holleczek B, Rossi S, Domenic A, Innos K, Minicozzi P, Francisci S, Hackl M, Eisemann N, Brenner H, EUROCARE-5 Working Group (2015) On-going improvement and persistent differences in the survival for patients with colon and rectum cancer across Europe 1999–2007–Results from the EUROCARE-5 study. Eur J Cancer 51:2158–2168. https://doi.org/10.1016/j.ejca.2015.07.024
Hosseini A, Minucci S (2017) A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics 9:1123–1142. https://doi.org/10.2217/epi-2017-0022
Huang Z, Li S, Song W, Li X, Li Q, Zhang Z, Han Y, Zhang X, Miao S, Du R, Wang L (2013) Lysine-specific demethylase 1 (LSD1/KDM1A) contributes to colorectal tumorigenesis via activation of the Wnt/β-catenin pathway by down-regulating Dickkopf-1 (DKK1). PloS One 8:e70077. https://doi.org/10.1371/journal.pone.0070077
Jiang T, Wang Y, Zhou F, Gao G, Ren S, Zhou C (2016) Prognostic value of high EZH2 expression in patients with different types of cancer: a systematic review with meta-analysis. Oncotarget 7:4584–4597. https://doi.org/10.18632/oncotarget.6612
Jie D, Zhongmin Z, Guoqing L, Sheng L, Yi Z, Jing W, Liang Z (2013) Positive expression of LSD1 and negative expression of E-cadherin correlate with metastasis and poor prognosis of colon cancer. Dig Dis Sci 58:1581–1589. https://doi.org/10.1007/s10620-012-2552-2
Jin L, Hanigan CL, Wu Y, Wang W, Park BH, Woster PM, Casero RA (2013) Loss of LSD1 (lysine-specific demethylase 1) suppresses growth and alters gene expression of human colon cancer cells in a p53-and DNMT1 (DNA methyltransferase 1)-independent manner. Biochem J 449:459–468. https://doi.org/10.1042/BJ20121360
Jones PA, Issa JP, Baylin S (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17:630–641. https://doi.org/10.1038/nrg.2016.93
Kim HA, Koo BK, Cho JH, Kim YY, Seong J, Chang HJ, Oh YM, Stange DE, Park JG, Hwang D, Kong YY (2012) Notch1 counteracts WNT/β-catenin signaling through chromatin modification in colorectal cancer. J Clin Invest 122:3248–3259. https://doi.org/10.1172/JCI61216
Kurihara H, Maruyama R, Ishiguro K, Kanno S, Yamamoto I, Ishigami K, Mitsuhashi K, Igarashi H, Ito M, Tanuma T, Sukawa Y, Okita K, Hasegawa T, Imai K, Yamamoto H, Shinomura Y, Nosho K (2016) The relationship between EZH2 expression and microRNA-31 in colorectal cancer and the role in evolution of the serrated pathway. Oncotarget 7:12704–12717. https://doi.org/10.18632/oncotarget.7260
Liu YL, Gao X, Jiang Y, Zhang G, Sun ZC, Cui BB, Yang YM (2015) Expression and clinicopathological significance of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J Cancer Res Clin Oncol 141:661–669. https://doi.org/10.1007/s00432-014-1854-5
Lu H, Li G, Zhou C, Jin W, Qian X, Wang Z, Pan H, Jin H, Wang X (2016) Regulation and role of post-translational modifications of enhancer of zeste homologue 2 in cancer development. Am J Cancer Res 6:2737–2754
Meng X, Huang Z, Wang R, Jiao Y, Li H, Xu X, Feng R, Zhu K, Jiang S, Yan H, Yu J (2014) The prognostic role of EZH2 expression in rectal cancer patients treated with neoadjuvant chemoradiotherapy. Radiat Oncol 9:188. https://doi.org/10.1186/1748-717X-9-188
Mimori K, Ogawa K, Okamoto M, Sudo T, Inoue H, Mori M (2005) Clinical significance of enhancer of zeste homolog 2 expression in colorectal cancer cases. Eur J Surg Oncol 31:376–380. https://doi.org/10.1016/j.ejso.2004.11.001
Olcina MM, Leszczynska KB, Senra JM, Isa NF, Harada H, Hammond EM (2016) H3K9me3 facilitates hypoxia-induced p53-dependent apoptosis through repression of APAK. Oncogene 35:793–799. https://doi.org/10.1038/onc.2015.134
Puccini A, Berger MD, Naseem M, Tokunaga R, Battaglin F, Cao S, Hanna DL, McSkane M, Soni S, Zhang W, Lenz HJ (2017) Colorectal cancer: Epigenetic alterations and their clinical implications. Biochim Biophys Acta 1868:439–448. https://doi.org/10.1016/j.bbcan.2017.09.003
Robbez-Masson L, Tie CH, Rowe HM (2017) Cancer cells, on your histone marks, get SETDB1, silence retrotransposons, and go! J Cell Biol 216:3429–3431. https://doi.org/10.1083/jcb.201710068
Schmoll HJ, Van Cutsem E, Stein A, Valentini V, Glimelius B, Haustermans K, Nordlinger B, van de Velde CJ, Balmana J, Regula J, Nagtegaal ID, Beets-Tan RG, Arnold D, Ciardiello F, Hoff P, Kerr D, Kohne CH, Labianca R, Price T, Scheithauer W, Sobrero A, Tabernero J, Aderka D, Barroso S, Bodoky G, Douillard JY, El Ghazaly H, Gallardo J, Garin A, Glynne-Jones R, Jordan K, Meshceryakov A, Papamichail D, Pfeiffer P, Souglakos I, Turhal S, Cervantes A (2012) ESMO Consensus Guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol 23:2479–2516. https://doi.org/10.1093/annonc/mds236
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67:177–193. https://doi.org/10.3322/caac.21395
Takawa M, Masuda K, Kunizaki M, Daigo Y, Takagi K, Iwai Y, Cho HS, Toyokawa G, Yamane Y, Maejima K, Field HI, Kobayashi T, Akasu T, Sugiyama M, Tsuchiya E, Atomi Y, Ponder BA, Nakamura Y, Hamamoto R (2011) Validation of the histone methyltransferase EZH2 as a therapeutic target for various types of human cancer and as a prognostic marker. Cancer Sci 102:1298–1305. https://doi.org/10.1111/j.1349-7006.2011.01958.x
Tamagawa H, Oshima T, Numata M, Yamamoto N, Shiozawa M, Morinaga S, Nakamura Y, Yoshihara M, Sakuma Y, Kameda Y, Akaike M, Yukawa N, Rino Y, Masusa M, Miyagi Y (2013) Global histone modification of H3K27 correlates with the outcomes in patients with metachronous liver metastasis of colorectal cancer. Eur J Surg Oncol 39:655–661. https://doi.org/10.1016/j.ejso.2013.02.023
Tan J, Yang X, Jiang X, Zhou J, Li Z, Lee PL, Li B, Robson P, Yu Q (2014) Integrative epigenome analysis identifies a Polycomb-targeted differentiation program as a tumor-suppressor event epigenetically inactivated in colorectal cancer. Cell Death Dis 5:e1324. https://doi.org/10.1038/cddis.2014.283
Vilorio-Marqués L, Martín V, Diez-Tascón C, González-Sevilla MF, Fernández-Villa T, Honrado E, Davila-Batista V, Molina AJ (2017) The role of EZH2 in overall survival of colorectal cancer: a meta-analysis. Sci Rep 7:13806. https://doi.org/10.1038/s41598-017-13670-z
Wang CG, Ye YJ, Yuan J, Liu FF, Zhang H, Wang S (2010) EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol 16:2421–2427. https://doi.org/10.3748/wjg.v16.i19.2421
Wassef M, Margueron R (2016) The multiple facets of PRC2 alterations in cancers. J Mol Biol 429:1978–1993. https://doi.org/10.1016/j.jmb.2016.10.012
Yokoyama Y, Hieda M, Nishioka Y, Matsumoto A, Higashi S, Kimura H, Yamamoto H, Mori M, Matsuura S, Matsuura N (2013) Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci 104:889–895. https://doi.org/10.1111/cas.12166
Funding
This study was funded by Research Center of Portuguese Oncology Institute of Porto (Grant number PI 74-CI-IPOP-19-2016) and by European Community’s Seventh Framework Programme—Grant number FP7-HEALTH-F5-2009-241783.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Carmen Jerónimo and Rui Henrique jointly served as senior authors
Rights and permissions
About this article
Cite this article
Carvalho, S., Freitas, M., Antunes, L. et al. Prognostic value of histone marks H3K27me3 and H3K9me3 and modifying enzymes EZH2, SETDB1 and LSD-1 in colorectal cancer. J Cancer Res Clin Oncol 144, 2127–2137 (2018). https://doi.org/10.1007/s00432-018-2733-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-018-2733-2