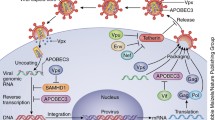
Abstract
SAMHD1 was initially described for its ability to efficiently restrict HIV-1 replication in myeloid cells and resting CD4+ T cells. However, a growing body of evidence suggests that SAMHD1-mediated restriction is by far not limited to lentiviruses, but seems to be a general concept that applies to most retroviruses and at least a number of DNA viruses. SAMHD1 anti-viral activity was long believed to be solely due to its ability to deplete cellular dNTPs by enzymatic degradation. However, since its discovery, several new functions have been attributed to SAMHD1. It has been demonstrated to bind nucleic acids, to modulate innate immunity, as well as to participate in the DNA damage response and resolution of stalled replication forks. Consequently, it is likely that SAMHD1-mediated anti-viral activity is not or not exclusively mediated through its dNTPase activity. Therefore, in this review, we summarize current knowledge on SAMHD1 cellular functions and systematically discuss how these functions could contribute to the restriction of a broad range of viruses besides retroviruses: herpesviruses, poxviruses and hepatitis B virus. Furthermore, we aim to highlight different ways how viruses counteract SAMHD1-mediated restriction to bypass the SAMHD1-mediated block to viral infection.

Similar content being viewed by others
References
Goldstone DC, Ennis-Adeniran V, Hedden JJ et al (2011) HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature 480(7377):379–382. https://doi.org/10.1038/nature10623
Lahouassa H, Daddacha W, Hofmann H et al (2012) SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13(3):223–228. https://doi.org/10.1038/ni.2236
Chen S, Bonifati S, Qin Z et al (2018) SAMHD1 suppresses innate immune responses to viral infections and inflammatory stimuli by inhibiting the NF-κB and interferon pathways. Proc Natl Acad Sci USA 115(16):E3798–E3807. https://doi.org/10.1073/pnas.1801213115
Daddacha W, Koyen AE, Bastien AJ et al (2017) SAMHD1 promotes DNA end resection to facilitate DNA repair by homologous recombination. Cell Rep 20(8):1921–1935. https://doi.org/10.1016/j.celrep.2017.08.008
Coquel F, Silva M-J, Técher H et al (2018) SAMHD1 acts at stalled replication forks to prevent interferon induction. Nature 557(7703):57–61. https://doi.org/10.1038/s41586-018-0050-1
Beloglazova N, Flick R, Tchigvintsev A et al (2013) Nuclease activity of the human SAMHD1 protein implicated in the Aicardi-Goutieres syndrome and HIV-1 restriction. J Biol Chem 288(12):8101–8110. https://doi.org/10.1074/jbc.M112.431148
Goncalves A, Karayel E, Rice GI et al (2012) SAMHD1 is a nucleic-acid binding protein that is mislocalized due to aicardi-goutières syndrome-associated mutations. Hum Mutat 33(7):1116–1122. https://doi.org/10.1002/humu.22087
Seamon KJ, Sun Z, Shlyakhtenko LS et al (2015) SAMHD1 is a single-stranded nucleic acid binding protein with no active site-associated nuclease activity. Nucleic Acids Res 43(13):6486–6499. https://doi.org/10.1093/nar/gkv633
Tüngler V, Staroske W, Kind B et al (2013) Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med 91(6):759–770. https://doi.org/10.1007/s00109-013-0995-3
Rice GI, Bond J, Asipu A et al (2009) Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet 41(7):829–832. https://doi.org/10.1038/ng.373
Crow YJ, Livingston JH (2008) Aicardi-Goutières syndrome: an important Mendelian mimic of congenital infection. Dev Med Child Neurol 50(6):410–416. https://doi.org/10.1111/j.1469-8749.2008.02062.x
Rentoft M, Lindell K, Tran P et al (2016) Heterozygous colon cancer-associated mutations of SAMHD1 have functional significance. Proc Natl Acad Sci USA 113(17):4723–4728. https://doi.org/10.1073/pnas.1519128113
Clifford R, Louis T, Robbe P et al (2014) SAMHD1 is mutated recurrently in chronic lymphocytic leukemia and is involved in response to DNA damage. Blood 123(7):1021–1031. https://doi.org/10.1182/blood-2013-04-490847
Powell RD, Holland PJ, Hollis T et al (2011) Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem 286(51):43596–43600. https://doi.org/10.1074/jbc.C111.317628
Franzolin E, Pontarin G, Rampazzo C et al (2013) The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci USA 110(35):14272–14277. https://doi.org/10.1073/pnas.1312033110
Seamon KJ, Bumpus NN, Stivers JT (2016) Single-stranded nucleic acids bind to the tetramer interface of SAMHD1 and prevent formation of the catalytic homotetramer. Biochemistry 55(44):6087–6099. https://doi.org/10.1021/acs.biochem.6b00986
White TE, Brandariz-Nuñez A, Valle-Casuso JC et al (2013) Contribution of SAM and HD domains to retroviral restriction mediated by human SAMHD1. Virology 436(1):81–90. https://doi.org/10.1016/j.virol.2012.10.029
Ryoo J, Choi J, Oh C et al (2014) The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med 20(8):936–941. https://doi.org/10.1038/nm.3626
Choi J, Ryoo J, Oh C et al (2015) SAMHD1 specifically restricts retroviruses through its RNase activity. Retrovirology 12:46. https://doi.org/10.1186/s12977-015-0174-4
Antonucci JM, St Gelais C, Silva S de et al (2016) SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med 22(10):1072–1074. https://doi.org/10.1038/nm.4163
Laguette N, Sobhian B, Casartelli N et al (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474(7353):654–657. https://doi.org/10.1038/nature10117
Lenzi GM, Domaoal RA, Kim D-H et al (2014) Kinetic variations between reverse transcriptases of viral protein X coding and noncoding lentiviruses. Retrovirology 11:111. https://doi.org/10.1186/s12977-014-0111-y
Riess M, Fuchs NV, Idica A et al (2017) Interferons induce expression of SAMHD1 in monocytes through down-regulation of miR-181a and miR-30a. J Biol Chem 292(1):264–277. https://doi.org/10.1074/jbc.M116.752584
Yang S, Zhan Y, Zhou Y et al (2016) Interferon regulatory factor 3 is a key regulation factor for inducing the expression of SAMHD1 in antiviral innate immunity. Sci Rep 6:29665. https://doi.org/10.1038/srep29665
Silva S de, Hoy H, Hake TS et al (2013) Promoter methylation regulates SAMHD1 gene expression in human CD4+ T cells. J Biol Chem 288(13):9284–9292. https://doi.org/10.1074/jbc.M112.447201
Cohen D, Adamovich Y, Reuven N et al (2010) Hepatitis B virus activates deoxynucleotide synthesis in nondividing hepatocytes by targeting the R2 gene. Hepatology 51(5):1538–1546. https://doi.org/10.1002/hep.23519
Ricardo-Lax I, Ramanan V, Michailidis E et al (2015) Hepatitis B virus induces RNR-R2 expression via DNA damage response activation. J Hepatol 63(4):789–796. https://doi.org/10.1016/j.jhep.2015.05.017
Cameron JM, McDougall I, Marsden HS et al (1988) Ribonucleotide reductase encoded by herpes simplex virus is a determinant of the pathogenicity of the virus in mice and a valid antiviral target. J Gen Virol 69(Pt 10):2607–2612. https://doi.org/10.1099/0022-1317-69-10-2607
Whitehurst CB, Ning S, Bentz GL et al (2009) The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J Virol 83(9):4345–4353. https://doi.org/10.1128/JVI.02195-08
Lembo D, Gribaudo G, Hofer A et al (2000) Expression of an altered ribonucleotide reductase activity associated with the replication of murine cytomegalovirus in quiescent fibroblasts. J Virol 74(24):11557–11565. https://doi.org/10.1128/JVI.74.24.11557-11565.2000
Gammon DB, Gowrishankar B, Duraffour S et al (2010) Vaccinia virus-encoded ribonucleotide reductase subunits are differentially required for replication and pathogenesis. PLoS Pathog 6(7):e1000984. https://doi.org/10.1371/journal.ppat.1000984
Buller RM, Smith GL, Cremer K et al (1985) Decreased virulence of recombinant vaccinia virus expression vectors is associated with a thymidine kinase-negative phenotype. Nature 317(6040):813–815
Field HJ, Wildy P (1978) The pathogenicity of thymidine kinase-deficient mutants of herpes simplex virus in mice. J Hyg 81(02):267. https://doi.org/10.1017/S0022172400025109
Cribier A, Descours B, Valadão ALC et al (2013) Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3(4):1036–1043. https://doi.org/10.1016/j.celrep.2013.03.017
White TE, Brandariz-Nuñez A, Valle-Casuso JC et al (2013) The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13(4):441–451. https://doi.org/10.1016/j.chom.2013.03.005
Welbourn S, Dutta SM, Semmes OJ et al (2013) Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol 87(21):11516–11524. https://doi.org/10.1128/JVI.01642-13
Sommer AFR, Rivière L, Qu B et al (2016) Restrictive influence of SAMHD1 on Hepatitis B Virus life cycle. Sci Rep 6:26616. https://doi.org/10.1038/srep26616
Hu J, Qiao M, Chen Y et al (2018) Cyclin E2-CDK2 mediates SAMHD1 phosphorylation to abrogate its restriction of HBV replication in hepatoma cells. FEBS Lett 592(11):1893–1904. https://doi.org/10.1002/1873-3468.13105
Schott K, Fuchs NV, Derua R et al (2018) Dephosphorylation of the HIV-1 restriction factor SAMHD1 is mediated by PP2A-B55α holoenzymes during mitotic exit. Nat Commun 9(1):2227. https://doi.org/10.1038/s41467-018-04671-1
Zhang K, Lv DW, Li R (2018) Conserved herpesvirus protein kinases target SAMHD1 to facilitate virus replication. Cell Rep. https://doi.org/10.2139/ssrn.3255560
Hansen EC, Seamon KJ, Cravens SL et al (2014) GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci USA 111(18):E1843–E1851. https://doi.org/10.1073/pnas.1401706111
Mauney CH, Rogers LC, Harris RS et al (2017) The SAMHD1 dNTP triphosphohydrolase is controlled by a redox switch. Antioxid Redox Signal 27(16):1317–1331. https://doi.org/10.1089/ars.2016.6888
Wang Z, Bhattacharya A, White T et al (2018) Functionality of redox-active cysteines is required for restriction of retroviral replication by SAMHD1. Cell Rep 24(4):815–823. https://doi.org/10.1016/j.celrep.2018.06.090
Kyei GB, Cheng X, Ramani R et al (2015) Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17(1):98–106. https://doi.org/10.1016/j.chom.2014.11.009
Ji X, Tang C, Zhao Q et al (2014) Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci USA 111(41):E4305–E4314. https://doi.org/10.1073/pnas.1412289111
Yan J, Kaur S, DeLucia M et al (2013) Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem 288(15):10406–10417. https://doi.org/10.1074/jbc.M112.443796
Amie SM, Bambara RA, Kim B (2013) GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J Biol Chem 288(35):25001–25006. https://doi.org/10.1074/jbc.C113.493619
Kennedy EM, Gavegnano C, Nguyen L et al (2010) Ribonucleoside triphosphates as substrate of human immunodeficiency virus type 1 reverse transcriptase in human macrophages. J Biol Chem 285(50):39380–39391. https://doi.org/10.1074/jbc.M110.178582
Baldauf H-M, Pan X, Erikson E et al (2012) SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18(11):1682–1687. https://doi.org/10.1038/nm.2964
Welbourn S, Strebel K (2016) Low dNTP levels are necessary but may not be sufficient for lentiviral restriction by SAMHD1. Virology 488:271–277. https://doi.org/10.1016/j.virol.2015.11.022
Koç A, Wheeler LJ, Mathews CK et al (2004) Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. J Biol Chem 279(1):223–230. https://doi.org/10.1074/jbc.M303952200
Arnold LH, Groom HCT, Kunzelmann S et al (2015) Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog 11(10):e1005194. https://doi.org/10.1371/journal.ppat.1005194
Bhattacharya A, Wang Z, White T et al (2016) Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Sci Rep 6:31353. https://doi.org/10.1038/srep31353
Yan J, Hao C, DeLucia M et al (2015) CyclinA2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J Biol Chem 290(21):13279–13292. https://doi.org/10.1074/jbc.M115.646588
Kretschmer S, Wolf C, König N et al (2015) SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 74(3):e17. https://doi.org/10.1136/annrheumdis-2013-204845
Rice GI, Meyzer C, Bouazza N et al (2018) Reverse-transcriptase inhibitors in the Aicardi–Goutières syndrome. N Engl J Med 379(23):2275–2277. https://doi.org/10.1056/NEJMc1810983
Mathews CK (2015) Deoxyribonucleotide metabolism, mutagenesis and cancer. Nat Rev Cancer 15(9):528–539. https://doi.org/10.1038/nrc3981
Ganai RA, Johansson E (2016) DNA replication—a matter of fidelity. Mol Cell 62(5):745–755. https://doi.org/10.1016/j.molcel.2016.05.003
Mathews CK (2006) DNA precursor metabolism and genomic stability. FASEB J 20(9):1300–1314. https://doi.org/10.1096/fj.06-5730rev
Takahashi A, Loo TM, Okada R et al (2018) Downregulation of cytoplasmic DNases is implicated in cytoplasmic DNA accumulation and SASP in senescent cells. Nat Commun 9(1):1249. https://doi.org/10.1038/s41467-018-03555-8
Gluck S, Guey B, Gulen MF et al (2017) Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat Cell Biol 19(9):1061–1070. https://doi.org/10.1038/ncb3586
Yang H, Wang H, Ren J et al (2017) cGAS is essential for cellular senescence. Proc Natl Acad Sci USA 114(23):E4612–E4620. https://doi.org/10.1073/pnas.1705499114
Maelfait J, Bridgeman A, Benlahrech A et al (2016) Restriction by SAMHD1 limits cGAS/STING-dependent innate and adaptive immune responses to HIV-1. Cell Rep 16(6):1492–1501. https://doi.org/10.1016/j.celrep.2016.07.002
Cotta-Ramusino C, Fachinetti D, Lucca C et al (2005) Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell 17(1):153–159. https://doi.org/10.1016/j.molcel.2004.11.032
Quinet A, Lemaçon D, Vindigni A (2017) Replication fork reversal: players and guardians. Mol Cell 68(5):830–833. https://doi.org/10.1016/j.molcel.2017.11.022
Patel DR, Weiss RS (2018) A tough row to hoe: when replication forks encounter DNA damage. Biochem Soc Trans 46(6):1643–1651. https://doi.org/10.1042/BST20180308
Ranjha L, Howard SM, Cejka P (2018) Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma 127(2):187–214. https://doi.org/10.1007/s00412-017-0658-1
Symington LS, Gautier J (2011) Double-strand break end resection and repair pathway choice. Annu Rev Genet 45:247–271. https://doi.org/10.1146/annurev-genet-110410-132435
Liu T, Huang J (2016) DNA end resection: facts and mechanisms. Genom Proteom Bioinform 14(3):126–130. https://doi.org/10.1016/j.gpb.2016.05.002
Bhattacharya S, Srinivasan K, Abdisalaam S et al (2017) RAD51 interconnects between DNA replication, DNA repair and immunity. Nucleic Acids Res 45(8):4590–4605. https://doi.org/10.1093/nar/gkx126
Wolf C, Rapp A, Berndt N et al (2016) RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat Commun 7:11752. https://doi.org/10.1038/ncomms11752
Hartlova A, Erttmann SF, Am Raffi F et al (2015) DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42(2):332–343. https://doi.org/10.1016/j.immuni.2015.01.012
Li T, Chen ZJ (2018) The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med 215(5):1287–1299. https://doi.org/10.1084/jem.20180139
Descours B, Cribier A, Chable-Bessia C et al (2012) SAMHD1 restricts HIV-1 reverse transcription in quiescent CD4(+) T-cells. Retrovirology 9:87. https://doi.org/10.1186/1742-4690-9-87
Schott K, Riess M, König R (2017) Role of innate genes in HIV replication. Curr Top Microbiol Immunol. https://doi.org/10.1007/82_2017_29
Sáez-Cirión A, Manel N (2018) Immune responses to retroviruses. Annu Rev Immunol 36:193–220. https://doi.org/10.1146/annurev-immunol-051116-052155
Honeycutt JB, Wahl A, Baker C et al (2016) Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest 126(4):1353–1366. https://doi.org/10.1172/JCI84456
Honeycutt JB, Thayer WO, Baker CE et al (2017) HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23(5):638–643. https://doi.org/10.1038/nm.4319
Rodrigues V, Ruffin N, San-Roman M et al (2017) Myeloid cell interaction with HIV: a complex relationship. Front Immunol 8:1698. https://doi.org/10.3389/fimmu.2017.01698
Bloch N, O’Brien M, Norton TD et al (2014) HIV type 1 infection of plasmacytoid and myeloid dendritic cells is restricted by high levels of SAMHD1 and cannot be counteracted by Vpx. AIDS Res Hum Retrovirus 30(2):195–203. https://doi.org/10.1089/AID.2013.0119
Berger A, Sommer AFR, Zwarg J et al (2011) SAMHD1-deficient CD14 + cells from individuals with Aicardi-Goutières syndrome are highly susceptible to HIV-1 infection. PLoS Pathog 7(12):e1002425. https://doi.org/10.1371/journal.ppat.1002425
Hrecka K, Hao C, Gierszewska M et al (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474(7353):658–661. https://doi.org/10.1038/nature10195
Goujon C, Riviere L, Jarrosson-Wuilleme L et al (2007) SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. https://doi.org/10.1186/1742-4690-4-2
Srivastava S, Swanson SK, Manel N et al (2008) Lentiviral Vpx accessory factor targets VprBP/DCAF1 substrate Adaptor for Cullin 4 E3 ubiquitin ligase to enable macrophage infection. PLoS Pathog. https://doi.org/10.1371/journal.ppat.1000059
Krupovic M, Blomberg J, Coffin JM et al (2018) Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol. https://doi.org/10.1128/JVI.00515-18
Gao WY, Cara A, Gallo RC et al (1993) Low levels of deoxynucleotides in peripheral blood lymphocytes: A strategy to inhibit human immunodeficiency virus type 1 replication. Proc Natl Acad Sci USA 90(19):8925–8928
Korin YD, Zack JA (1999) Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J Virol 73(8):6526–6532
Plesa G, Dai J, Baytop C et al (2007) Addition of deoxynucleosides enhances human immunodeficiency virus type 1 integration and 2LTR formation in resting CD4+ T cells. J Virol 81(24):13938–13942. https://doi.org/10.1128/JVI.01745-07
Kim B, Nguyen LA, Daddacha W et al (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem 287(26):21570–21574. https://doi.org/10.1074/jbc.C112.374843
Ruiz A, Pauls E, Badia R et al (2015) Cyclin D3-dependent control of the dNTP pool and HIV-1 replication in human macrophages. Cell Cycle 14(11):1657–1665. https://doi.org/10.1080/15384101.2015.1030558
Mlcochova P, Sutherland KA, Watters SA et al (2017) A G1-like state allows HIV-1 to bypass SAMHD1 restriction in macrophages. EMBO J 36(5):604–616. https://doi.org/10.15252/embj.201696025
Lee EJ, Seo JH, Park J-H et al (2017) SAMHD1 acetylation enhances its deoxynucleotide triphosphohydrolase activity and promotes cancer cell proliferation. Oncotarget 8(40):68517–68529. https://doi.org/10.18632/oncotarget.19704
Lamoliatte F, Caron D, Durette C et al (2014) Large-scale analysis of lysine SUMOylation by SUMO remnant immunoaffinity profiling. Nat Commun 5:5409. https://doi.org/10.1038/ncomms6409
Hendriks IA, Lyon D, Young C et al (2017) Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 24(3):325–336. https://doi.org/10.1038/nsmb.3366
Lumpkin RJ, Gu H, Zhu Y et al (2017) Site-specific identification and quantitation of endogenous SUMO modifications under native conditions. Nat Commun 8(1):1171. https://doi.org/10.1038/s41467-017-01271-3
Elia AEH, Boardman AP, Wang DC et al (2015) Quantitative proteomic atlas of ubiquitination and acetylation in the DNA damage response. Mol Cell 59(5):867–881. https://doi.org/10.1016/j.molcel.2015.05.006
Hofmann H, Norton TD, Schultz ML et al (2013) Inhibition of CUL4A Neddylation causes a reversible block to SAMHD1-mediated restriction of HIV-1. J Virol 87(21):11741–11750. https://doi.org/10.1128/JVI.02002-13
Campbell EM, Hope TJ (2015) HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat Rev Microbiol 13(8):471–483. https://doi.org/10.1038/nrmicro3503
Bejarano DA, Puertas MC, Börner K et al (2018) Detailed characterization of early HIV-1 replication dynamics in primary human macrophages. Viruses. https://doi.org/10.3390/v10110620
Brandariz-Nuñez A, Valle-Casuso JC, White TE et al (2012) Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. https://doi.org/10.1186/1742-4690-9-49
Le Grice SFJ (2012) Human immunodeficiency virus reverse transcriptase: 25 years of research, drug discovery, and promise. J Biol Chem 287(49):40850–40857. https://doi.org/10.1074/jbc.R112.389056
Hughes SH (2015) Reverse transcription of retroviruses and LTR retrotransposons. Microbiol Spectr 3(2):MDNA3-0027-2014. https://doi.org/10.1128/microbiolspec.MDNA3-0027-2014
Meng X, Zhao X (2017) Replication fork regression and its regulation. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fow110
Arhel NJ, Souquere-Besse S, Munier S et al (2007) HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J 26(12):3025–3037. https://doi.org/10.1038/sj.emboj.7601740
Roshal M, Kim B, Zhu Y et al (2003) Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem 278(28):25879–25886. https://doi.org/10.1074/jbc.M303948200
Zimmerman ES, Sherman MP, Blackett JL et al (2006) Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol 80(21):10407–10418. https://doi.org/10.1128/JVI.01212-06
Laguette N, Brégnard C, Hue P et al (2014) Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156(1–2):134–145. https://doi.org/10.1016/j.cell.2013.12.011
Berger G, Lawrence M, Hué S et al (2015) G2/M cell cycle arrest correlates with primate lentiviral Vpr interaction with the SLX4 complex. J Virol 89(1):230–240. https://doi.org/10.1128/JVI.02307-14
Fregoso OI, Emerman M (2016) Activation of the DNA damage response is a conserved function of HIV-1 and HIV-2 Vpr that is independent of SLX4 recruitment. MBio. https://doi.org/10.1128/mBio.01433-16
Daniel R, Katz RA, Skalka AM (1999) A role for DNA-PK in retroviral DNA integration. Science 284(5414):644–647
Li L (2001) Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J 20(12):3272–3281. https://doi.org/10.1093/emboj/20.12.3272
Skalka AM, Katz RA (2005) Retroviral DNA integration and the DNA damage response. Cell Death Differ 12(Suppl 1):971–978. https://doi.org/10.1038/sj.cdd.4401573
Hu WS, Temin HM (1990) Retroviral recombination and reverse transcription. Science 250(4985):1227–1233
Levy DN, Aldrovandi GM, Kutsch O et al (2004) Dynamics of HIV-1 recombination in its natural target cells. Proc Natl Acad Sci USA 101(12):4204–4209. https://doi.org/10.1073/pnas.0306764101
Temin HM (1991) Sex and recombination in retroviruses. Trends Genet 7(3):71–74. https://doi.org/10.1016/0168-9525(91)90272-R
Rawson JMO, Nikolaitchik OA, Keele BF et al (2018) Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic Acids Res 46(20):10535–10545. https://doi.org/10.1093/nar/gky910
König R, Zhou Y, Elleder D et al (2008) Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135(1):49–60. https://doi.org/10.1016/j.cell.2008.07.032
Kumar D, Abdulovic AL, Viberg J et al (2011) Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res 39(4):1360–1371. https://doi.org/10.1093/nar/gkq829
Watt DL, Buckland RJ, Lujan SA et al (2016) Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools. Nucleic Acids Res 44(4):1669–1680. https://doi.org/10.1093/nar/gkv1298
Chabes A, Georgieva B, Domkin V et al (2003) Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112(3):391–401
Poli J, Tsaponina O, Crabbé L et al (2012) dNTP pools determine fork progression and origin usage under replication stress. EMBO J 31(4):883–894. https://doi.org/10.1038/emboj.2011.470
Buckland RJ, Watt DL, Chittoor B et al (2014) Increased and imbalanced dNTP pools symmetrically promote both leading and lagging strand replication infidelity. PLoS Genet 10(12):e1004846. https://doi.org/10.1371/journal.pgen.1004846
Bester AC, Roniger M, Oren YS et al (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145(3):435–446. https://doi.org/10.1016/j.cell.2011.03.044
Kennedy EM, Amie SM, Bambara RA et al (2012) Frequent incorporation of ribonucleotides during HIV-1 reverse transcription and their attenuated repair in macrophages. J Biol Chem 287(17):14280–14288. https://doi.org/10.1074/jbc.M112.348482
Operario DJ, Balakrishnan M, Bambara RA et al (2006) Reduced dNTP interaction of human immunodeficiency virus type 1 reverse transcriptase promotes strand transfer. J Biol Chem 281(43):32113–32121. https://doi.org/10.1074/jbc.M604665200
Laguette N, Rahm N, Sobhian B et al (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11(2):205–217. https://doi.org/10.1016/j.chom.2012.01.007
Lim ES, Fregoso OI, McCoy CO et al (2012) The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11(2):194–204. https://doi.org/10.1016/j.chom.2012.01.004
Gramberg T, Kahle T, Bloch N et al (2013) Restriction of diverse retroviruses by SAMHD1. Retrovirology 10:26. https://doi.org/10.1186/1742-4690-10-26
Mereby SA, Maehigashi T, Holler JM et al (2018) Interplay of ancestral non-primate lentiviruses with the virus-restricting SAMHD1 proteins of their hosts. J Biol Chem 293(42):16402–16412. https://doi.org/10.1074/jbc.RA118.004567
Kaushik R, Zhu X, Stranska R et al (2009) A cellular restriction dictates the permissivity of nondividing monocytes/macrophages to lentivirus and gammaretrovirus infection. Cell Host Microbe 6(1):68–80. https://doi.org/10.1016/j.chom.2009.05.022
Sze A, Belgnaoui SM, Olagnier D et al (2013) Host restriction factor SAMHD1 limits human T cell leukemia virus type 1 infection of monocytes via STING-mediated apoptosis. Cell Host Microbe 14(4):422–434. https://doi.org/10.1016/j.chom.2013.09.009
Yan H, Li W (2015) Sodium taurocholate cotransporting polypeptide acts as a receptor for hepatitis B and D virus. Dig Dis 33(3):388–396. https://doi.org/10.1159/000371692
Beck J (2007) Hepatitis B virus replication. WJG 13(1):48. https://doi.org/10.3748/wjg.v13.i1.48
Gómez-Moreno A, Garaigorta U (2017) Hepatitis B virus and DNA damage response: interactions and consequences for the infection. Viruses. https://doi.org/10.3390/v9100304
Bock CT, Schwinn S, Locarnini S et al (2001) Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307(1):183–196. https://doi.org/10.1006/jmbi.2000.4481
Chen Z, Zhu M, Pan X et al (2014) Inhibition of Hepatitis B virus replication by SAMHD1. Biochem Biophys Res Commun 450(4):1462–1468. https://doi.org/10.1016/j.bbrc.2014.07.023
Jeong GU, Park I-H, Ahn K et al (2016) Inhibition of hepatitis B virus replication by a dNTPase-dependent function of the host restriction factor SAMHD1. Virology 495:71–78. https://doi.org/10.1016/j.virol.2016.05.001
Gearhart TL, Bouchard MJ (2010) The hepatitis B virus X protein modulates hepatocyte proliferation pathways to stimulate viral replication. J Virol 84(6):2675–2686. https://doi.org/10.1128/JVI.02196-09
Tuttleman JS, Pourcel C, Summers J (1986) Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47(3):451–460
Gaillard RK, Barnard J, Lopez V et al (2002) Kinetic analysis of wild-type and YMDD mutant hepatitis B virus polymerases and effects of deoxyribonucleotide concentrations on polymerase activity. Antimicrob Agents Chemother 46(4):1005–1013. https://doi.org/10.1128/AAC.46.4.1005-1013.2002
Wang H, Kim S, Ryu W-S (2009) DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J Virol 83(11):5815–5824. https://doi.org/10.1128/JVI.00011-09
Baumert TF, Rösler C, Malim MH et al (2007) Hepatitis B virus DNA is subject to extensive editing by the human deaminase APOBEC3C. Hepatology 46(3):682–689. https://doi.org/10.1002/hep.21733
Wu TT, Coates L, Aldrich CE et al (1990) In hepatocytes infected with duck hepatitis B virus, the template for viral RNA synthesis is amplified by an intracellular pathway. Virology 175(1):255–261
Kim ET, White TE, Brandariz-Núñez A et al (2013) SAMHD1 restricts herpes simplex virus 1 in macrophages by limiting DNA replication. J Virol 87(23):12949–12956. https://doi.org/10.1128/JVI.02291-13
Hollenbaugh JA, Gee P, Baker J et al (2013) Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog 9(6):e1003481. https://doi.org/10.1371/journal.ppat.1003481
Grosche L, Kummer M, Steinkasserer A (2017) What goes around, comes around—HSV-1 replication in monocyte-derived dendritic cells. Front Microbiol 8:2149. https://doi.org/10.3389/fmicb.2017.02149
Ellermann-Eriksen S (2005) Macrophages and cytokines in the early defence against herpes simplex virus. Virol J 2:59. https://doi.org/10.1186/1743-422X-2-59
Weller SK, Coen DM (2012) Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb Perspect Biol 4(9):a013011. https://doi.org/10.1101/cshperspect.a013011
Smith S, Weller SK (2015) HSV-I and the cellular DNA damage response. Future Virol 10(4):383–397. https://doi.org/10.2217/fvl.15.18
Fregoso OI, Ahn J, Wang C et al (2013) Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog 9(7):e1003496. https://doi.org/10.1371/journal.ppat.1003496
Sharp PM, Bailes E, Stevenson M et al (1996) Gene acquisition in HIV and SIV. Nature 383(6601):586–587. https://doi.org/10.1038/383586a0
Ahn J, Hao C, Yan J et al (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287(15):12550–12558. https://doi.org/10.1074/jbc.M112.340711
Schwefel D, Groom HCT, Boucherit VC et al (2014) Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505(7482):234–238. https://doi.org/10.1038/nature12815
Schwefel D, Boucherit VC, Christodoulou E et al (2015) Molecular determinants for recognition of divergent SAMHD1 proteins by the lentiviral accessory protein Vpx. Cell Host Microbe 17(4):489–499. https://doi.org/10.1016/j.chom.2015.03.004
Accola MA, Bukovsky AA, Jones MS et al (1999) A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J Virol 73(12):9992–9999
Sunseri N, O’Brien M, Bhardwaj N et al (2011) Human immunodeficiency virus type 1 modified to package Simian immunodeficiency virus Vpx efficiently infects macrophages and dendritic cells. J Virol 85(13):6263–6274. https://doi.org/10.1128/JVI.00346-11
Zhang C, Silva S de, Wang J-H et al (2012) Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS One 7(5):e37477. https://doi.org/10.1371/journal.pone.0037477
Nyamweya S, Hegedus A, Jaye A et al (2013) Comparing HIV-1 and HIV-2 infection: Lessons for viral immunopathogenesis. Rev Med Virol 23(4):221–240. https://doi.org/10.1002/rmv.1739
Jin C, Peng X, Liu F et al (2016) Interferon-induced sterile alpha motif and histidine/aspartic acid domain-containing protein 1 expression in astrocytes and microglia is mediated by microRNA-181a. AIDS 30(13):2053–2064. https://doi.org/10.1097/QAD.0000000000001166
Su B, Biedma ME, Lederle A et al (2014) Dendritic cell-lymphocyte cross talk downregulates host restriction factor SAMHD1 and stimulates HIV-1 replication in dendritic cells. J Virol 88(9):5109–5121. https://doi.org/10.1128/JVI.03057-13
Boyer PL, Clark PK, Hughes SH (2012) HIV-1 and HIV-2 reverse transcriptases: different mechanisms of resistance to nucleoside reverse transcriptase inhibitors. J Virol 86(10):5885–5894. https://doi.org/10.1128/JVI.06597-11
Chayama K, Suzuki Y, Kobayashi M et al (1998) Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology 27(6):1711–1716. https://doi.org/10.1002/hep.510270634
Goldstein DJ, Weller SK (1988) Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: Isolation and characterization of an ICP6 lacZ insertion mutant. J Virol 62(1):196–205
Jacobson JG, Leib DA, Goldstein DJ et al (1989) A herpes simplex virus ribonucleotide reductase deletion mutant is defective for productive acute and reactivatable latent infections of mice and for replication in mouse cells. Virology 173(1):276–283
Mineta T, Rabkin SD, Yazaki T et al (1995) Attenuated multi-mutated herpes simplex virus-1 for the treatment of malignant gliomas. Nat Med 1(9):938–943
Lembo D, Donalisio M, Hofer A et al (2004) The ribonucleotide reductase R1 homolog of murine cytomegalovirus is not a functional enzyme subunit but is required for pathogenesis. J Virol 78(8):4278–4288. https://doi.org/10.1128/JVI.78.8.4278-4288.2004
Kit S, Qavi H, Dubbs DR et al (1983) Attenuated marmoset herpesvirus isolated from recombinants of virulent marmoset herpesvirus and hybrid plasmids. J Med Virol 12(1):25–36
Rice AP, Kimata JT (2015) Subversion of cell cycle regulatory mechanisms by HIV. Cell Host Microbe 17(6):736–740. https://doi.org/10.1016/j.chom.2015.05.010
Jowett JB, Planelles V, Poon B et al (1995) The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 69(10):6304–6313
Romani B, Shaykh Baygloo N, Aghasadeghi MR et al (2015) HIV-1 Vpr protein enhances proteasomal degradation of MCM10 DNA replication factor through the Cul4-DDB1VprBP E3 ubiquitin ligase to induce G2/M cell cycle arrest. J Biol Chem 290(28):17380–17389. https://doi.org/10.1074/jbc.M115.641522
He J, Choe S, Walker R et al (1995) Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69(11):6705–6711
Sakai K, Dimas J, Lenardo MJ (2006) The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc Natl Acad Sci USA 103(9):3369–3374. https://doi.org/10.1073/pnas.0509417103
Wang J, Reuschel EL, Shackelford JM et al (2011) HIV-1 Vif promotes the G1- to S-phase cell-cycle transition. Blood 117(4):1260–1269. https://doi.org/10.1182/blood-2010-06-289215
Benn J, Schneider RJ (1995) Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci 92(24):11215–11219. https://doi.org/10.1073/pnas.92.24.11215
Lee YI, Kang-Park S, Do SI (2001) The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J Biol Chem 276(20):16969–16977. https://doi.org/10.1074/jbc.M011263200
Bouchard M, Giannakopoulos S, Wang EH et al (2001) Hepatitis B virus HBx protein activation of cyclin A-cyclin-dependent kinase 2 complexes and G1 transit via a Src kinase pathway. J Virol 75(9):4247–4257. https://doi.org/10.1128/JVI.75.9.4247-4257.2001
Mukherji A, Janbandhu VC, Kumar V (2007) HBx-dependent cell cycle deregulation involves interaction with cyclin E/A-cdk2 complex and destabilization of p27Kip1. Biochem J 401(1):247–256. https://doi.org/10.1042/BJ20061091
Spector DH (2015) Human cytomegalovirus riding the cell cycle. Med Microbiol Immunol 204(3):409–419. https://doi.org/10.1007/s00430-015-0396-z
Kuny CV, Chinchilla K, Culbertson MR et al (2010) Cyclin-dependent kinase-like function is shared by the beta- and gamma-subset of the conserved herpesvirus protein kinases. PLoS Pathog 6(9):e1001092. https://doi.org/10.1371/journal.ppat.1001092
Li R, Liao G, Nirujogi RS et al (2015) Phosphoproteomic profiling reveals Epstein–Barr virus protein kinase integration of DNA damage response and mitotic signaling. PLoS Pathog 11(12):e1005346. https://doi.org/10.1371/journal.ppat.1005346
Posch W, Steger M, Knackmuss U et al (2015) Complement-opsonized HIV-1 overcomes restriction in dendritic cells. PLoS Pathog 11(6):e1005005. https://doi.org/10.1371/journal.ppat.1005005
Ayinde D, Bruel T, Cardinaud S et al (2015) SAMHD1 limits HIV-1 antigen presentation by monocyte-derived dendritic cells. J Virol 89(14):6994–7006. https://doi.org/10.1128/JVI.00069-15
Etienne L, Hahn BH, Sharp PM et al (2013) Gene loss and adaptation to hominids underlie the ancient origin of HIV-1. Cell Host Microbe 14(1):85–92. https://doi.org/10.1016/j.chom.2013.06.002
Thientosapol ES, Bosnjak D, Durack T et al (2018) SAMHD1 enhances immunoglobulin hypermutation by promoting transversion mutation. Proc Natl Acad Sci USA 115(19):4921–4926. https://doi.org/10.1073/pnas.1719771115
Acknowledgements
We would like to thank Kerstin Schott and Lise Lauterbach-Rivière for critical comments on the manuscript. This study was supported by the Deutsche Forschungsgemeinschaft (DFG; CRC1292 Project TP04 to R.K. and SPP1923 Project KO 4573/1-1 to R.K.) and by Deutsches Zentrum für Infektionsforschung (HZI2016Z10).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Edited by: Matthias J. Reddehase.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Special Issue on Immunological Imprinting during Chronic Viral Infection.
Rights and permissions
About this article
Cite this article
Majer, C., Schüssler, J.M. & König, R. Intertwined: SAMHD1 cellular functions, restriction, and viral evasion strategies. Med Microbiol Immunol 208, 513–529 (2019). https://doi.org/10.1007/s00430-019-00593-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-019-00593-x