Abstract
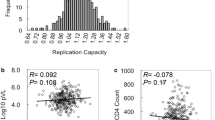
In a Nairobi-Kenyan cohort of 50 HIV-1 positive patients, we analysed the prevalence of HIV-1 subtypes and human leucocyte antigen (HLA) alleles. From this cohort, 33 patients were selected for the analysis of HIV-1 infection progression markers (i.e. CD4 cell counts and viral loads) and their association with HIV-1 genetic variability and subtype, and patient’s HLA type. HIV-1 gag genetic variability, analysed using bioinformatics tools, showed an inverse relationship with CD4 cell count whereas with viral load that relationship was direct. Certain HLA types and viral subtypes were also found to associate with patients’ viral load. Associations between disease parameters and the genetic makeup of the host and virus may be crucial in determining the outcome of HIV-1 infection.

Similar content being viewed by others
References
Hewitt EW (2003) The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110(2):163–169
Arien KK, Vanham G, Arts EJ (2007) Is HIV-1 evolving to a less virulent form in humans? Nat Rev Microbiol 5(2):141–151
Haynes BF, Pantaleo G, Fauci AS (1996) Toward an understanding of the correlates of protective immunity to HIV infection. Science 271(5247):324–328
Kaur G, Mehra N (2009) Genetic determinants of HIV-1 infection and progression to AIDS: immune response genes. Tissue Antigens 74(5):373–385
Khoja S, Ojwang P, Khan S, Okinda N, Harania R, Ali S (2008) Genetic analysis of HIV-1 subtypes in Nairobi, Kenya. PLoS One 3(9):e3191
Reekie J, Gatell JM, Yust I, Bakowska E, Rakhmanova A, Losso M et al (2011) Fatal and nonfatal AIDS and non-AIDS events in HIV-1-positive individuals with high CD4 cell counts according to viral load strata. AIDS 25(18):2259–2268
Bettinotti MP, Mitsuishi Y, Bibee K, Lau M, Terasaki PI (1997) Comprehensive method for the typing of HLA-A, B, and C alleles by direct sequencing of PCR products obtained from genomic DNA. J Immunother 20(6):425–430
Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, Marsh SG (2011) The IMGT/HLA database. Nucleic Acids Res 39(Database issue):D1171–D1176
Rose PP, Korber BT (2000) Detecting hypermutations in viral sequences with an emphasis on G → A hypermutation. Bioinformatics 16(4):400–401
Yang OO (2009) Candidate vaccine sequences to represent intra- and inter-clade HIV-1 variation. PLoS One 4(10):e7388
Cao K, Moormann AM, Lyke KE, Masaberg C, Sumba OP, Doumbo OK et al (2004) Differentiation between African populations is evidenced by the diversity of alleles and haplotypes of HLA class I loci. Tissue Antigens 63(4):293–325
Bird TG, Kaul R, Rostron T, Kimani J, Embree J, Dunn PP et al (2002) HLA typing in a Kenyan cohort identifies novel class I alleles that restrict cytotoxic T-cell responses to local HIV-1 clades. AIDS 16(14):1899–1904
Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD et al (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330(6010):1551–1557
Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ et al (1999) HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283(5408):1748–1752
Tang J, Tang S, Lobashevsky E, Myracle AD, Fideli U, Aldrovandi G et al (2002) Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J Virol 76(16):8276–8284
Trachtenberg E, Korber B, Sollars C, Kepler TB, Hraber PT, Hayes E et al (2003) Advantage of rare HLA supertype in HIV disease progression. Nat Med 9(7):928–935
Kiwanuka N, Robb M, Laeyendecker O, Kigozi G, Wabwire-Mangen F, Makumbi FE et al (2009) HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J Acquir Immune Defic Syndr 54(2):180–184
Baeten JM, Chohan B, Lavreys L, Chohan V, McClelland RS, Certain L et al (2007) HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis 195(8):1177–1180
Acknowledgments
For this study, Syed Hani Abidi’s training was funded by HIV Research Trust (Scholarship No. HIVRT 10-059).
Conflict of interest
Authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abidi, S.H., Shahid, A., Lakhani, L.S. et al. HIV-1 progression links with viral genetic variability and subtype, and patient’s HLA type: analysis of a Nairobi-Kenyan cohort. Med Microbiol Immunol 203, 57–63 (2014). https://doi.org/10.1007/s00430-013-0314-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-013-0314-1