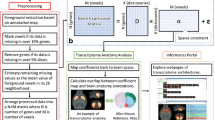
Abstract
Gene coexpression patterns carry rich information regarding enormously complex brain structures and functions. Characterization of these patterns in an unbiased, integrated, and anatomically comprehensive manner will illuminate the higher-order transcriptome organization and offer genetic foundations of functional circuitry. Here using dictionary learning and sparse coding, we derived coexpression networks from the space-resolved anatomical comprehensive in situ hybridization data from Allen Mouse Brain Atlas dataset. The key idea is that if two genes use the same dictionary to represent their original signals, then their gene expressions must share similar patterns, thereby considering them as “coexpressed.” For each network, we have simultaneous knowledge of spatial distributions, the genes in the network and the extent a particular gene conforms to the coexpression pattern. Gene ontologies and the comparisons with published gene lists reveal biologically identified coexpression networks, some of which correspond to major cell types, biological pathways, and/or anatomical regions.







Similar content being viewed by others
References
Allocco DJ, Kohane IS, Butte AJ (2004) Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinform 5:18. doi:10.1186/1471-2105-5-18
Bando SY, Silva FN, Costa LDF, Silva AV, Pimentel-Silva LR, Castro LH et al (2013) Complex network analysis of CA3 transcriptome reveals pathogenic and compensatory pathways in refractory temporal lobe epilepsy. PLoS One 8(11):e79913. doi:10.1371/journal.pone.0079913
Bernard A, Lubbers LS, Tanis KQ, Luo R, Podtelezhnikov AA, Finney EM et al (2012) Transcriptional architecture of the primate neocortex. Neuron 73(6):1083–1099. doi:10.1016/j.neuron.2012.03.002.Transcriptional
Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW (2004) Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci USA 101(7):2173–2178. doi:10.1073/pnas.0308512100
Bohland JW, Bokil H, Pathak SD, Lee C-K, Ng L, Lau C et al (2010) Clustering of spatial gene expression patterns in the mouse brain and comparison with classical neuroanatomy. Methods 50(2):105–112. doi:10.1016/j.ymeth.2009.09.001
Brown CD, Johnson DS, Sidow A (2007) Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science 317(September):1557–1560
Cahoy J, Emery B, Kaushal A, Foo L, Zamanian J, Christopherson K et al (2004) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neuronsci 28(1):264–278. doi:10.1523/JNEUROSCI.4178-07.2008
Carter H, Hofree M, Ideker T (2013) Genotype to phenotype via network analysis. Curr Opin Genet Dev 23(6):611–621. doi:10.1016/j.gde.2013.10.003
Chen H, Li K, Zhu D, Jiang X, Yuan Y, Lv P et al (2013) Inferring group-wise consistent multimodal brain networks via multi-view spectral clustering. IEEE Trans Med Imaging 32(9):1576–1586. doi:10.1109/TMI.2013.2259248
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(5):P3. doi:10.1186/gb-2003-4-5-p3
Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S et al (2009) Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol 10(5):R55. doi:10.1186/gb-2009-10-5-r55
Dong S, Li C, Wu P, Tsien JZ, Hu Y (2007) Environment enrichment rescues the neurodegenerative phenotypes in presenilins-deficient mice. Eur J Neurosci 26(1):101–112. doi:10.1111/j.1460-9568.2007.05641.x
Efron B, Hastie T, Johnstone I, Tibshirani R (2004) Least angle regression. Ann Stat 32(2):407–499
Eisen MB, Spellman PT, Brown PO, Botstein D (1999) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95(22):12930–12933. doi:10.1073/pnas.95.25.14863
Gaiteri C, Ding Y, French B, Tseng GC, Sibille E (2014) Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav 13(1):13–24. doi:10.1111/gbb.12106
Ge H, Liu Z, Church GM, Vidal M (2001) Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 29(4):482–486. doi:10.1038/ng776
Grange P, Bohland JW, Okaty BW, Sugino K, Bokil H, Nelson SB et al (2014) Cell-type-based model explaining coexpression patterns of genes in the brain. Proc Natl Acad Sci USA 111(14):5397–5402. doi:10.1073/pnas.1312098111
Hawrylycz M, Bernard A, Lau C, Sunkin SM, Chakravarty MM, Lein ES et al (2010) Areal and laminar differentiation in the mouse neocortex using large scale gene expression data. Methods 50(2):113–121. doi:10.1016/j.ymeth.2009.09.005
Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL et al (2015) Canonical genetic signatures of the adult human brain. Nat Neurosci. doi:10.1038/nn.4171
Jiang CH, Tsien JZ, Schultz PG, Hu Y (2001) The effects of aging on gene expression in the hypothalamus and cortex of mice. PNAS 98(4):1930–1934. doi:10.1073/pnas.98.4.1930
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinform 9:559. doi:10.1186/1471-2105-9-559
Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P (2004) Coexpression analysis of human genes across many microarray data sets. Genome Res 14:1085–1094. doi:10.1101/gr.1910904.1
Lein ES, Zhao X, Gage FH (2004) Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci 24(15):3879–3889. doi:10.1523/JNEUROSCI.4710-03.2004
Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A et al (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445(7124):168–176. doi:10.1038/nature05453
Lu T, Pan Y, Kao S-Y, Li C, Kohane I, Chan J, Yankner BA (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429(June):883–891. doi:10.1038/nature02618.1
Luxburg U (2007) A tutorial on spectral clustering. Stat Comput 17(4):395–416. doi:10.1007/s11222-007-9033-z
Mairal J, Elad M, Sapiro G (2008) Sparse representation for color image restoration. IEEE Trans Image Process 17(1):53–69. doi:10.1109/TIP.2007.911828
Mairal J, Bach F, Ponce J, Sapiro G (2010) Online learning for matrix factorization and sparse coding. J Mach Learn Res 11:19–60. http://portal.acm.org/citation.cfm?id=1756008
Miao H, Crabb AW, Hernandez MR, Lukas TJ (2010) Modulation of factors affecting optic nerve head astrocyte migration. Invest Ophthalmol Vis Sci 51(8):4096–4103. doi:10.1167/iovs.10-5177
Miller J (2014) Transcriptional landscape of the prenatal human brain. Nature 508(7495):199–206. doi:10.1038/nature13185.Transcriptional
Miller JA, Horvath S, Geschwind DH (2010) Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA 107(28):12698–12703. doi:10.1073/pnas.0914257107
Miller JA, Cai C, Langfelder P, Geschwind DH, Kurian SM, Salomon DR, Horvath S (2011) Strategies for aggregating gene expression data: the collapseRows R function. BMC Bioinform 12(1):322. doi:10.1186/1471-2105-12-322
Mody M, Cao Y, Cui Z, Tay KY, Shyong A, Shimizu E et al (2001) Genome-wide gene expression profiles of the developing mouse hippocampus. PNAS 98:8862–8867. doi:10.1073/pnas.141244998
Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD (2007) Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci 8(6):427–437. doi:10.1038/nrn2151
Ng L, Pathak SD, Kuan C, Lau C, Dong H, Sodt A et al (2007) Neuroinformatics for genome-wide 3D gene expression mapping in the mouse brain. IEEE/ACM Trans Comput Biol Bioinf 4(3):382–392. doi:10.1109/TCBB.2007.1035
Ng L, Bernard A, Lau C, Overly CC, Dong H-W, Kuan C et al (2009) An anatomic gene expression atlas of the adult mouse brain. Nat Neurosci 12(3):356–362. doi:10.1038/nn.2281
O’Leary DD, Chou SJ, Sahara S (2007) Area patterning of the mammalian cortex. Neuron 56(2):252–269. doi:10.1016/j.neuron.2007.10.010
Oldham MC, Horvath S, Geschwind DH (2006) Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA 103(47):17973–17978. doi:10.1073/pnas.0605938103
Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH (2008) Functional organization of the transcriptome in human brain. Nat Neurosci 11(11):1271–1282. doi:10.1038/nn.2207
Oldham MC, Langfelder P, Horvath S (2012) Network methods for describing sample relationships in genomic datasets: application to Huntington’s disease. BMC Syst Biol 6(1):63. doi:10.1186/1752-0509-6-63
Olshausen BA, Field DJ (2004) Sparse coding of sensory inputs. Curr Opin Neurobiol 14(4):481–487. doi:10.1016/j.conb.2004.07.007
Peng H, Long F, Zhou J, Leung G, Eisen MB, Myers EW (2007) Automatic image analysis for gene expression patterns of fly embryos. BMC Cell Biol 8(Suppl 1):S7. doi:10.1186/1471-2121-8-S1-S7
Quinones-Hinojosa A, Chaichana K (2007) The human subventricular zone: a source of new cells and a potential source of brain tumors. Exp Neurol 205(2):313–324. doi:10.1016/j.expneurol.2007.03.016
Rampon C, Jiang CH, Dong H, Tang YP, Lockhart DJ, Schultz PG et al (2000) Effects of environmental enrichment on gene expression in the brain. Proc Natl Acad Sci USA 97(23):12880–12884. doi:10.1073/pnas.97.23.12880
Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302(5643):249–255. doi:10.1126/science.1087447
Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C et al (2006) Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci 9(1):99–107. doi:10.1038/nn1618
Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E et al (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 96(6):2907–2912. doi:10.1073/pnas.96.6.2907
Tavazoie S, Hughes JD, Campbell MJ, Cho RJ, Church GM (1999) Systematic determination of genetic network architecture. Nat Genet 22(3):281–285. doi:10.1038/10343
Winden KD, Oldham MC, Mirnics K, Ebert PJ, Swan CH, Levitt P et al (2009) The organization of the transcriptional network in specific neuronal classes. Mol Syst Biol 5(291):291. doi:10.1038/msb.2009.46
Wright E, Ng L, Guillozet-Bongarts A (2007) Annotation report on cerebellar cortex, pukinje cell layer. http://community.brain-map.org/download/attachments/798/cbxpu.pdf?version=1
Acknowledgements
T. Liu is supported by NIH R01 DA-033393, NSF CAREER Award IIS-1149260, NIH R01 AG-042599, NSF BME-1302089, NSF BCS-1439051 and NSF DBI-1564736.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Chen, H., Jiang, X. et al. Discover mouse gene coexpression landscapes using dictionary learning and sparse coding. Brain Struct Funct 222, 4253–4270 (2017). https://doi.org/10.1007/s00429-017-1460-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00429-017-1460-9