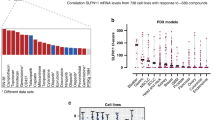
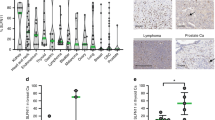
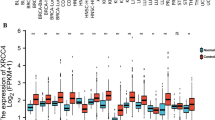
Abstract
DNA-damaging agents include first-line drugs such as platinum (cisplatin, carboplatin), topoisomerase inhibitors (etoposide, doxorubicin), and replication inhibitors (cytarabine, gemcitabine). Despite their wide and long usage, there is no clinically available biomarker to predict responses to these drugs. Schlafen 11 (SLFN11), a putative DNA/RNA helicase, recently emerged as a dominant determinant of sensitivity to these drugs by enforcing the replication block in response to DNA damage. Since the clinical importance of SLFN11 is implicated, a comprehensive analysis of SLFN11 expression across human organs will provide a practical resource to develop the utility of SLFN11 in the clinic. In this study, we established a scoring system of SLFN11 expression by immunohistochemistry (IHC) and assessed SLFN11 expression in ~ 700 malignant as well as the adjacent non-tumor tissues across 16 major human adult organs. We found that the SLFN11 expression is tissue specific and varies during tumorigenesis. Although The Cancer Genome Atlas (TCGA) is a prevailing tool to assess gene expression in various malignant and normal tissues, our IHC data exhibited obvious discrepancy from the TCGA data in several organs. Importantly, SLFN11-negative tumors, potentially non-responders to DNA-damaging agents, were largely overrated in TCGA because TCGA samples are a mixture of infiltrating immune cells, including T cells, B cells, and macrophages, which have strong SLFN11 expression. Thus, our study reveals the significance of immunohistochemical procedures for evaluating expression of SLFN11 in patient samples and provides a robust resource of SLFN11 expression across adult human organs.





Similar content being viewed by others
References
Allison Stewart C, Tong P, Cardnell RJ, Sen T, Li L, Gay CM, Masrorpour F, Fan Y, Bara RO, Feng Y, Ru Y, Fujimoto J, Kundu ST, Post LE, Yu K, Shen Y, Glisson BS, Wistuba I, Heymach JV, Gibbons DL, Wang J, Byers LA (2017) Dynamic variations in epithelial-to-mesenchymal transition (EMT), ATM, and SLFN11 govern response to PARP inhibitors and cisplatin in small cell lung cancer. Oncotarget 8:28575. https://doi.org/10.18632/oncotarget.15338
Ballestrero A, Bedognetti D, Ferraioli D, Franceschelli P, Labidi-Galy SI, Leo E, Murai J, Pommier Y, Tsantoulis P, Vellone VG, Zoppoli G (2017) Report on the first SLFN11 monothematic workshop: from function to role as a biomarker in cancer. J Transl Med 15:199. https://doi.org/10.1186/s12967-017-1296-3
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA (2012) The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483:603–607. https://doi.org/10.1038/nature11003
Deng Y, Cai Y, Huang Y, Yang Z, Bai Y, Liu Y, Deng X, Wang J (2015) High SLFN11 expression predicts better survival for patients with KRAS exon 2 wild type colorectal cancer after treated with adjuvant oxaliplatin-based treatment. BMC Cancer 15:833. https://doi.org/10.1186/s12885-015-1840-6
Gardner EE, Lok BH, Schneeberger VE, Desmeules P, Miles LA, Arnold PK, Ni A, Khodos I, de Stanchina E, Nguyen T, Sage J, Campbell JE, Ribich S, Rekhtman N, Dowlati A, Massion PP, Rudin CM, Poirier JT (2017) Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 31:286–299. https://doi.org/10.1016/j.ccell.2017.01.006
Hao Y, Yan M, Heath BR, Lei YL, Xie Y (2019) Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput Biol 15:e1006976. https://doi.org/10.1371/journal.pcbi.1006976
He T, Zhang M, Zheng R, Zheng S, Linghu E, Herman JG, Guo M (2017) Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 9:849–862. https://doi.org/10.2217/epi-2017-0019
Isnaldi E, Ferraioli D, Ferrando L, Brohee S, Ferrando F, Fregatti P, Bedognetti D, Ballestrero A, Zoppoli G (2019) Schlafen-11 expression is associated with immune signatures and basal-like phenotype in breast cancer. Breast Cancer Res Treat 177:335–343. https://doi.org/10.1007/s10549-019-05313-w
Kang MH, Wang J, Makena MR, Lee JS, Paz N, Hall CP, Song MM, Calderon RI, Cruz RE, Hindle A, Ko W, Fitzgerald JB, Drummond DC, Triche TJ, Reynolds CP (2015) Activity of MM-398, nanoliposomal irinotecan (nal-IRI), in Ewing's family tumor xenografts is associated with high exposure of tumor to drug and high SLFN11 expression. Clin Cancer Res 21:1139–1150. https://doi.org/10.1158/1078-0432.CCR-14-1882
Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M, Jones TE, Landry S, Pan T, Weitzman MD, David M (2012) Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 491:125–128. https://doi.org/10.1038/nature11433
Lok BH, Gardner EE, Schneeberger VE, Ni A, Desmeules P, Rekhtman N, de Stanchina E, Teicher BA, Riaz N, Powell SN, Poirier JT, Rudin CM (2017) PARP inhibitor activity correlates with SLFN11 expression and demonstrates synergy with temozolomide in small cell lung cancer. Clin Cancer Res 23:523–535. https://doi.org/10.1158/1078-0432.CCR-16-1040
Magnuson AM, Kiner E, Ergun A, Park JS, Asinovski N, Ortiz-Lopez A, Kilcoyne A, Paoluzzi-Tomada E, Weissleder R, Mathis D, Benoist C (2018) Identification and validation of a tumor-infiltrating Treg transcriptional signature conserved across species and tumor types. Proc Natl Acad Sci U S A 115:E10672–E10681. https://doi.org/10.1073/pnas.1810580115
Marzi L, Szabova L, Gordon M, Weaver Ohler Z, Sharan SK, Beshiri ML, Etemadi M, Murai J, Kelly K, Pommier Y (2019) The indenoisoquinoline TOP1 inhibitors selectively target homologous recombination deficient- and Schlafen 11-positive cancer cells and synergize with olaparib. Clin Cancer Res. https://doi.org/10.1158/1078-0432.CCR-19-0419
Mezzadra R, de Bruijn M, Jae LT, Gomez-Eerland R, Duursma A, Scheeren FA, Brummelkamp TR, Schumacher TN (2019) SLFN11 can sensitize tumor cells towards IFN-gamma-mediated T cell killing. PLoS One 14:e0212053. https://doi.org/10.1371/journal.pone.0212053
Murai J, Feng Y, Yu GK, Ru Y, Tang SW, Shen Y, Pommier Y (2016) Resistance to PARP inhibitors by SLFN11 inactivation can be overcome by ATR inhibition. Oncotarget 7:76534–76550. https://doi.org/10.18632/oncotarget.12266
Murai J, Tang SW, Leo E, Baechler SA, Redon CE, Zhang H, Al Abo M, Rajapakse VN, Nakamura E, Jenkins LMM, Aladjem MI, Pommier Y (2018) SLFN11 blocks stressed replication forks independently of ATR. Mol Cell 69:371–384.e376. https://doi.org/10.1016/j.molcel.2018.01.012
Nogales V, Reinhold WC, Varma S, Martinez-Cardus A, Moutinho C, Moran S, Heyn H, Sebio A, Barnadas A, Pommier Y, Esteller M (2016) Epigenetic inactivation of the putative DNA/RNA helicase SLFN11 in human cancer confers resistance to platinum drugs. Oncotarget 7:3084–3097. https://doi.org/10.18632/oncotarget.6413
Rajapakse VN, Luna A, Yamade M, Loman L, Varma S, Sunshine M, Iorio F, Sousa FG, Elloumi F, Aladjem MI, Thomas A, Sander C, Kohn KW, Benes CH, Garnett M, Reinhold WC, Pommier Y (2018) CellMinerCDB for integrative cross-database genomics and pharmacogenomics analyses of cancer cell lines. iScience 10:247–264. https://doi.org/10.1016/j.isci.2018.11.029
Sakamoto N, Naito Y, Oue N, Sentani K, Uraoka N, Oo HZ, Yanagihara K, Aoyagi K, Sasaki H, Yasui W (2014) MicroRNA-148a is downregulated in gastric cancer, targets MMP7, and indicates tumor invasiveness and poor prognosis. Cancer Sci 105:236–243. https://doi.org/10.1111/cas.12330
Tang SW, Bilke S, Cao L, Murai J, Sousa FG, Yamade M, Rajapakse V, Varma S, Helman LJ, Khan J, Meltzer PS, Pommier Y (2015) SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing sarcoma. Clin Cancer Res 21:4184–4193. https://doi.org/10.1158/1078-0432.CCR-14-2112
Tang SW, Thomas A, Murai J, Trepel JB, Bates SE, Rajapakse VN, Pommier Y (2018) Overcoming resistance to DNA-targeted agents by epigenetic activation of Schlafen 11 (SLFN11) expression with class I histone deacetylase inhibitors. Clin Cancer Res 24:1944–1953. https://doi.org/10.1158/1078-0432.CCR-17-0443
Tian L, Song S, Liu X, Wang Y, Xu X, Hu Y, Xu J (2014) Schlafen-11 sensitizes colorectal carcinoma cells to irinotecan. Anti-Cancer Drugs 25:1175–1181. https://doi.org/10.1097/cad.0000000000000151
Zoppoli G, Regairaz M, Leo E, Reinhold WC, Varma S, Ballestrero A, Doroshow JH, Pommier Y (2012) Putative DNA/RNA helicase Schlafen-11 (SLFN11) sensitizes cancer cells to DNA-damaging agents. Proc Natl Acad Sci U S A 109:15030–15035. https://doi.org/10.1073/pnas.1205943109
Acknowledgments
We thank Mr. Shinichi Norimura for his technical assistance. This work was carried out with kind cooperation from the Research Center for Molecular Medicine of the Faculty of Medicine of Hiroshima University. We also thank the Analysis Center of Life Science of Hiroshima University for the use of their facilities.
Funding
This work was supported by Grants-in-Aid for Scientific Research (JP15H04713 and JP16K08691 to W.Y., JP16H06999 to N.S.) and (19H03505 to J.M.), Challenging Exploratory Research (26670175, JP16K15247 to W.Y.) from the Japan Society for the Promotion of Science, AMED (Japan Agency for Medical Research and Development) Project for Cancer Research and Therapeutic Evolution (to J.M.), and a research grant from The Uehara Memorial Foundation (to J.M.). Y.P. and V.R. are supported by the Center for Cancer Research, the Intramural program of the US National Cancer Institute, NIH (Z01 BC 006150).
Author information
Authors and Affiliations
Contributions
N.S., J.K., and Y.P. designed the study. T.T., D.T., and K.K. provided the patients’ clinical information. T.T., D.T., R.H., S.U., R.M., and V.N.R. performed the experiments and acquired the data. N.S., J.K., Y.P., and W.Y. interpreted the results. T.T., N.S., J.M., V.N.R., Y.P., and W.Y. drafted and edited the manuscript. All the authors read and approved the manuscript and agree to be accountable for all aspects of the research and in ensuring that the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval
This study was approved by the Ethics Committee of Kure Medical Center and Chugoku Cancer Center (Kure, Japan, no. 2019-36) and conformed to the ethical guidelines of the Declaration of Helsinki.
Consent to participate
All samples were obtained with patient consent.
Code availability
Statistical differences were evaluated using the Pearson’s test. Statistical analyses were conducted primarily using GraphPad Prism software (GraphPad Software Inc.).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Quality in Pathology
Rights and permissions
About this article
Cite this article
Takashima, T., Sakamoto, N., Murai, J. et al. Immunohistochemical analysis of SLFN11 expression uncovers potential non-responders to DNA-damaging agents overlooked by tissue RNA-seq. Virchows Arch 478, 569–579 (2021). https://doi.org/10.1007/s00428-020-02840-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00428-020-02840-6