Abstract
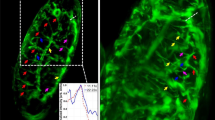
Methods to present three-dimensional (3D) and time series of 3D datasets (4D) are demonstrated using the recent advances in confocal microscopy and computer visualization. The process of cell sorting during tip formation in the slime mould Dictyostelium discoideum is examined as an example by in vivo confocal microscopy of spectrally different green fluorescent protein (GFP) variants as reporters of cell-type specific gene expression. Also, cell sorting of the co-aggregating slime mould species D. discoideum and D. mucoroides is observed using a GFP variant and a spectrally distinguishable fluorescent vital stain. The confocal data are handled as 3D and 4D datasets, their processing and the advantages of different methods of visualization are discussed step by step. Selected sequences of the experiments can be viewed on the Internet, giving a much better impression of the complex cellular movements during Dictyostelium morphogenesis than printed photographs.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 17 February 1998 / Accepted: 14 June 1998
Rights and permissions
About this article
Cite this article
Zimmermann, T., Siegert, F. 4D confocal microscopy of Dictyostelium discoideum morphogenesis and its presentation on the Internet. Dev Gene Evol 208, 411–420 (1998). https://doi.org/10.1007/s004270050198
Issue Date:
DOI: https://doi.org/10.1007/s004270050198